CDKN2A deletion is associated with immune desertification in diffuse pleural mesothelioma
- PMID: 40877968
- PMCID: PMC12392524
- DOI: 10.1186/s13046-025-03522-4
CDKN2A deletion is associated with immune desertification in diffuse pleural mesothelioma
Abstract
Introduction: Diffuse Pleural Mesothelioma (DPM) is a rare and incurable cancer. Immune checkpoint inhibitors (ICIs) marked some advances but only for a limited fraction of patients. Improving response prediction to ICIs is currently a clinical need in DPM. Deletion of CDKN2A gene, in chr9p21.3, is one of the most frequent alterations in DPM. As in other settings, deletion of CDKN2A locus has been associated with an immunosuppressive phenotype. Here we investigated the consequences of CDKN2A deletion (CDKN2Adel) on the tridimensional organization and function of immune infiltrate in DPM.
Methods: A retrospective cohort of 89 DPMs was analyzed and assessed for CDKN2Adel through digital droplet PCR. Immune-profiling was assessed by analyzing 770 immune-related genes by digital profiling. Finally, morphologically resolved, high-dimensional transcriptomic approach was used to reconstruct the spatial architecture of immune-tumor interaction in wild-type and deleted FFPE samples.
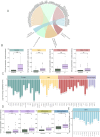
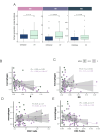
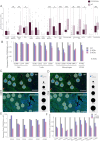
Results: CDKN2Adel was detected in 41.5% of DPMs and was associated with reduced survival (p = 0.04). Bulk gene expression identified 373 differentially expressed genes, of which 98.6% were downregulated in CDKN2Adel samples. These genes were enriched in several immune categories, suggesting significant immune deprivation in deleted tumors. Deconvolution analysis confirmed a major depletion of infiltrating immune cells including effector populations. Spatial transcriptomics revealed that this immunosuppressive phenotype was different according to histotype and prominent in the sarcomatoid lesions.
Conclusion: These data demonstrated that CDKN2Adel deeply affects the spatial organization of immune microenvironment by depleting immune-signaling and reducing or preventing immune infiltration, supporting the potential implementation of this alteration as ICIs predictive biomarker in DPM.
Keywords: CDKN2A; Diffuse pleural mesothelioma; Genetic alteration; Immunotherapy; Tumor immune microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the AVEN ethical committee (131/2023/TESS/IRCCSRE – DIMPLE). Consent for publication: Not Applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Janes SM, Alrifai D, Fennell DA. Perspectives on the treatment of malignant pleural mesothelioma. N Engl J Med. 2021;385:1207–18. - PubMed
-
- Giotti B, Dolasia K, Zhao W, Cai P, Sweeney R, Merritt E, Kiner E, Kim GS, Bhagwat A, Nguyen T, et al. Single-Cell view of tumor microenvironment gradients in pleural mesothelioma. Cancer Discov. 2024;14:2262–78. - PubMed
-
- Mangiante L, Alcala N, Sexton-Oates A, Di Genova A, Gonzalez-Perez A, Khandekar A, Bergstrom EN, Kim J, Liu X, Blazquez-Encinas R, et al. Multiomic analysis of malignant pleural mesothelioma identifies molecular axes and specialized tumor profiles driving intertumor heterogeneity. Nat Genet. 2023;55:607–18. - PMC - PubMed
-
- Galateau Salle F, Le Stang N, Tirode F, Courtiol P, Nicholson AG, Tsao MS, Tazelaar HD, Churg A, Dacic S, Roggli V, et al. Comprehensive molecular and pathologic evaluation of transitional mesothelioma assisted by deep learning approach: A Multi-Institutional study of the international mesothelioma panel from the MESOPATH reference center. J Thorac Oncol. 2020;15:1037–53. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

