Reprogramming of Fatty Acid Metabolism via PPARα-Orchestrated FADS2 in Keratinocytes Modulates Skin Inflammation in Psoriasis
- PMID: 40878384
- PMCID: PMC12561275
- DOI: 10.1002/advs.202417049
Reprogramming of Fatty Acid Metabolism via PPARα-Orchestrated FADS2 in Keratinocytes Modulates Skin Inflammation in Psoriasis
Abstract
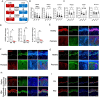
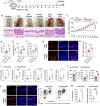
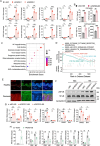
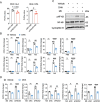
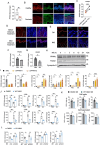
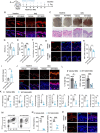
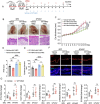
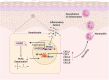
Psoriasis is a chronic inflammatory skin disorder characterized by keratinocyte hyper-proliferation and immune dysregulation. Recent evidence has implicated dysregulated polyunsaturated fatty acid (PUFA) metabolism in its pathogenesis. In this study, fatty acid desaturase 2 (FADS2), the rate-limiting Δ6-desaturase in PUFA biosynthesis, is identified as a central regulator of psoriatic inflammation. FADS2 expression is consistently reduced in keratinocytes from patients with psoriasis and in mouse models. Keratinocyte-intrinsic Fads2 knockdown exacerbates imiquimod-induced psoriasis-like dermatitis, which is marked by enhanced neutrophil recruitment and NF-κB activation, whereas Fads2 overexpression exerts protective effects and alleviates skin inflammation. In vitro, FADS2 knockdown in keratinocytes enhances M5-induced pro-inflammatory cytokine production, whereas FADS2 overexpression attenuates these effects. Lipidomic analysis reveals that impaired docosahexaenoic acid (DHA) biosynthesis is a key downstream consequence of FADS2 deficiency. Mechanistically, loss of FADS2 disrupts DHA biosynthesis, thus promoting an inflammatory response accompanied by increased NF-κB phosphorylation in keratinocytes to attract neutrophils. Furthermore, PPARα is identified as an upstream transcriptional activator of FADS2, and pharmacological activation of PPARα alleviates psoriatic inflammation in a FADS2-dependent manner. Together, these findings uncover a PPARα-FADS2-DHA-NF-κB axis that links lipid metabolism to immune regulation in psoriasis, highlighting a potential therapeutic strategy for restoring cutaneous immune homeostasis.
Keywords: FADS2; PPARα; fatty acid; inflammation; keratinocytes; psoriasis.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Griffiths C. E. M., Armstrong A. W., Gudjonsson J. E., Barker J. N. W. N., Lancet 2021, 397, 1301. - PubMed
-
- Kim J., Krueger J. G., Annu. Rev. Med. 2017, 68, 255. - PubMed
-
- Sarr O., Payne G. W., Hucik B., Abdelmagid S., Nakamura M. T., Ma D. W. L., Mutch D. M., J. Nutr. Biochem. 2019, 63, 140. - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFC2508106/National Key Research and Development Program of China
- 82273525:82430101:82273510:82273515:81900612:82103712/National Natural Science Foundation of China
- 2025GDZKZD06/Innovation Program of Shanghai Municipal Education Commission
- No.SHDC22022302/Clinical Research Plan of SHDC
- No. 2019KYQD08/Research Program of Shanghai Skin Disease Hospital
LinkOut - more resources
Full Text Sources
Medical
