Molecular Signatures and Functional Pathways of Human Monocytes and Macrophages in Allergy: An EAACI AllergoOncology Scoping Review
- PMID: 40878618
- PMCID: PMC12486325
- DOI: 10.1111/all.16672
Molecular Signatures and Functional Pathways of Human Monocytes and Macrophages in Allergy: An EAACI AllergoOncology Scoping Review
Abstract
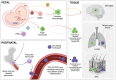
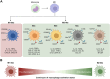
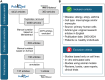
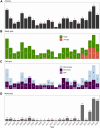
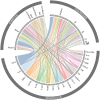
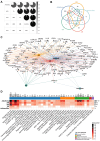
AllergoOncology explores the intersection of allergic diseases and cancer, focusing on shared immune mechanisms. While monocytes and macrophages are extensively studied in cancer, their roles in allergic diseases remain underexplored. To address this gap, we conducted a scoping review to systematically characterize the molecular landscape and related pathways of human monocytes and macrophages in allergy. An automated search of PubMed and Web of Science databases retrieved 4668 unique articles, which were manually curated based on predefined inclusion and exclusion criteria, yielding 138 eligible studies. From these, we identified 451 molecules associated with monocyte and macrophage responses across allergic disorders. Data analyses revealed a research bias towards blood-derived monocytes, underrepresentation of tissue-resident macrophages, and limited inclusion of miRNAs. Semantic similarity and pathway enrichment analyses highlighted a common molecular signature across major allergic disorders, with consistent enrichment in interleukin signaling and immune activation pathways. To enhance reproducibility and translational utility for researchers and clinicians, we developed ALO•HA, a web application for interactive data exploration. This overview of monocyte and macrophage molecular responses in human allergy underscores the need for integrative, human-focused approaches to better define their roles, and to guide future therapeutic strategies in allergic diseases and at the interface with oncology.
Keywords: AllergoOncology; allergy; data‐driven bioinformatics; macrophages; monocytes.
© 2025 The Author(s). Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
All authors have read and approved the manuscript. Any potential conflicts of interest related to the manuscript are listed here: S.N.K. is the founder and shareholder of Epsilogen Ltd. (formerly IGEM Therapeutics Ltd.) and has received funds from IGEM Therapeutics Ltd/Epsilogen Ltd. H.J.B., and M.G. are employed through a fund provided by Epsilogen Ltd. J.C. has been employed through a fund provided by Epsilogen Ltd. S.N.K. and H.J.B. are inventors of patents on antibody technologies. S.N.K. reports grants from Worldwide Cancer Research (24‐0087); Guy's and St Thomas's Foundation Trust Charity Melanoma Special Fund (573); the British Skin Foundation (006/R/22); the Biotechnology and Biological Sciences Research Council (BB/T008709/1); Cancer Research UK (C30122/A11527; C30122/A15774); the Cancer Research UK King's Health Partners Centre at King's College London (C604/A25135); the CRUK City of London Centre Award (C7893/A29290); Breast Cancer Now (147; KCL‐BCN‐Q3); the Medical Research Council (MR/L023091/1; MR/R015643/1; MR/V049445/1); Innovate UK (51746); S.N.K. has projects supported by the King's Health Partners Centre for Translational Medicine. The views expressed are those of the author(s) and not necessarily those of King's Health Partners. O.P. received research grants from MINECO, Ministerio de Ciencia e Innovación, CAM, Inmunotek S.L., Novartis, and AstraZeneca and fees for giving scientific lectures or participation in Advisory Boards from: AstraZeneca, Pfizer, GlaxoSmithKline, Inmunotek S.L., Novartis, Sanofi‐Genzyme and Regeneron. E.J.J. is inventor in EP2894478; “LCN2 as a tool for allergy diagnostic therapy”, EP 14150965.3, Year: 01/2014; US 14/204,570, immunoBON, owned by Biomedical International
Figures


References
-
- Turner M. C., Radzikowska U., Ferastraoaru D. E., et al., “AllergoOncology: Biomarkers and Refined Classification for Research in the Allergy and Glioma Nexus‐A Joint EAACI‐EANO Position Paper,” Allergy 79, no. 6 (2024): 1419–1439. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

