TIGER: A tdTomato in vivo genome-editing reporter mouse for investigating precision-editor delivery approaches
- PMID: 40880534
- PMCID: PMC12415246
- DOI: 10.1073/pnas.2506257122
TIGER: A tdTomato in vivo genome-editing reporter mouse for investigating precision-editor delivery approaches
Abstract
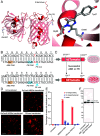
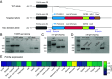
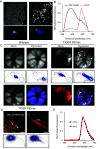
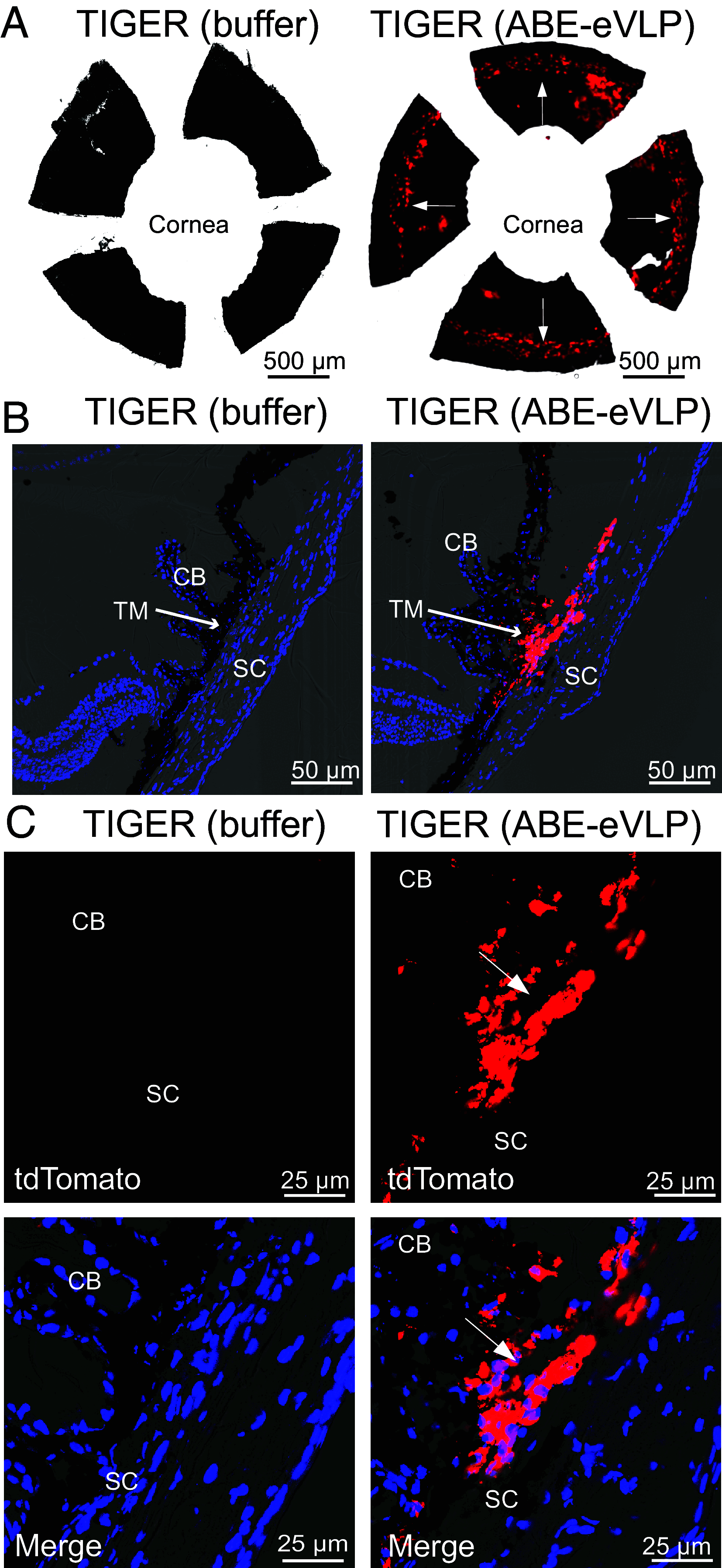
In vivo genome editing has the potential to address many inherited and environmental disorders. However, a major hurdle for the clinical translation of genome editing is safe, efficient delivery to disease-relevant tissues. A modality-agnostic reporter animal model that facilitates rapid, precise, and quantifiable assessment of functional delivery and editing could greatly enhance the evaluation and translation of delivery technologies. Here, we present the development of the tdTomato in vivo genome-editing reporter (TIGER) mouse, a reporter strain that harbors an integrated and constitutively expressed mutated tdTomato gene in the Polr2a locus. The mutations (Q115X, Q357X) abolish fluorescence, but successful adenine base editing (ABE) or prime editing (PE) restores tdTomato fluorescence. This mouse model facilitates the tissue- and cell type-specific assessment of genome editing agent delivery. We describe several editing strategies validated in vitro and demonstrate efficient ABE and PE in vivo using viral and nonviral delivery vectors targeting four cell types within the mouse eye: the retinal pigment epithelium, photoreceptors, Müller glia, and the trabecular meshwork. We show direct editing characterization in the ocular tissues via in vivo and ex vivo two-photon confocal microscopy and verify the spectral and fluorescence lifetime properties of tdTomato reporter in other mouse tissues. Additionally, we demonstrate successful adeno-associated virus (AAV)-mediated PE of extraocular tissues, including hepatocytes, skeletal muscle, and brain neurons by intravenous injection. Thus, the TIGER mouse facilitates the direct development, comparison, and optimization of delivery platforms for efficient and productive ABE or PE broadly applicable in vivo across multiple tissues tested in this study.
Keywords: CRISPR; base editing; genome editing; prime editing; reporter.
Conflict of interest statement
Competing interests statement:K.P. is a consultant for Polgenix Inc. and serves on the Scientific Advisory Board at Hyperion Eye Ltd. D.R.L. is a consultant, co-founder and/or equity owner of Beam Therapeutics, Prime Medicine, Pairwise Plants, and nChroma Bio, companies that use or deliver genome or epigenome editing agents. The Broad Institute has filed patent applications on base and prime editing. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- UG3 AI150551/AI/NIAID NIH HHS/United States
- R01 HD101642/HD/NICHD NIH HHS/United States
- R01 EY036994/EY/NEI NIH HHS/United States
- R01EY034501/HHS | NIH (NIH)
- RM1 HG009490/HG/NHGRI NIH HHS/United States
- R01HD101642/HHS | NIH (NIH)
- HHMI/HHMI (HHMI)
- P30EY034070/HHS | NIH (NIH)
- R01 EY026177/EY/NEI NIH HHS/United States
- Career-Starter Research Grants/Knights Templar Eye Foundation (KTEF)
- R01 EY034501/EY/NEI NIH HHS/United States
- N/A/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
- R35GM118062/HHS | NIH (NIH)
- P30 EY034070/EY/NEI NIH HHS/United States
- T32 GM008620/GM/NIGMS NIH HHS/United States
- F30 EY033642/EY/NEI NIH HHS/United States
- T32GM008620/HHS | NIH (NIH)
- R01EY036994/HHS | NIH (NIH)
- U01AI142756/HHS | NIH (NIH)
- RM1HG009490/HHS | NIH (NIH)
- F30EY033642/HHS | NIH (NIH)
- Unrestricted/Research to Prevent Blindness (RPB)
- R35 GM118062/GM/NIGMS NIH HHS/United States
- UG3AI150551/HHS | NIH (NIH)
- U01 AI142756/AI/NIAID NIH HHS/United States
- R01EY026177/HHS | NIH (NIH)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

