Cepharanthine hydrochloride inhibits prostate cancer progression by modulating gut microbiota and metabolites
- PMID: 40880650
- PMCID: PMC12380746
- DOI: 10.3389/fphar.2025.1627656
Cepharanthine hydrochloride inhibits prostate cancer progression by modulating gut microbiota and metabolites
Abstract
Background: Cepharanthine Hydrochloride (CH) is widely used in clinical settings to alleviate leukopenia caused by various tumors following radiotherapy and chemotherapy. However, it remains unclear whether CH have an inhibitory effect on the progression of prostate cancer, and whether this effect is mediated by gut microbiota. To address this question, the present study constructed normal mouse models of prostate cancer, as well as antibiotic-treated mouse models of prostate cancer.
Methods: CH were then administered via gavage to both groups of model mice. After treatment, the tumor sizes of the mice were measured, and feces, blood, and tumor tissues from both groups were collected for 16S rDNA, metabolomics, and transcriptomics sequencing analysis.
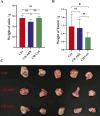
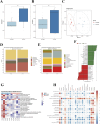
Results: Results showed CH treatment significantly suppressed prostate cancer growth in mice without antibiotic cocktail pretreatment, but not in antibiotic-pretreated mice. 16S rRNA sequencing revealed distinct gut microbiota alterations in CH-Ctrl versus Ctrl/CH-ABX groups, with increased g_Blautia, g_Lactobacillus, g_Butyricicoccus and decreased g_Akkermansia abundances. Metabolomic analysis identified 240 and 123 differentially abundant metabolites in CH-Ctrl vs Ctrl and CH-ABX, respectively. RNA-seq detected 579 and 530 differentially expressed genes in CH-Ctrl vs Ctrl and CH-ABX, respectively. Correlation analysis of differential gut microbiota, metabolites, and genes suggested that CH might inhibit prostate cancer growth by increasing the relative abundance of g_Blautia, g_Lactobacillus, and g_Butyricicoccus, suppressing g_Akkermansia proliferation, enhancing Acetylglycine metabolite production, upregulating Ttpa, Gm14964, Shc3, Elovl4 gene expression, and downregulating Gm10531, Bc021767 gene expression.
Conclusion: This study is the first to explore the potential mechanisms of gut microbiota-mediated CH treatment for prostate cancer, providing a scientific basis for the application of CH in PCa therapy.
Keywords: antibiotic cocktail; cepharanthine hydrochloride; gut microbiota; metabolites of gut microbiota; prostate cancer.
Copyright © 2025 Li, Luo, He, Dong, Jia, Sun, Zheng and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Gut microbiota depletion accelerates hematoma resolution and neurological recovery after intracerebral hemorrhage via p-coumaric acid-promoted Treg differentiation.Theranostics. 2025 Jun 9;15(14):6628-6650. doi: 10.7150/thno.113764. eCollection 2025. Theranostics. 2025. PMID: 40585999 Free PMC article.
-
Effects of a veterinary gastrointestinal diet on fecal characteristics, metabolites, and microbiota concentrations of adult cats treated with metronidazole.J Anim Sci. 2024 Jan 3;102:skae274. doi: 10.1093/jas/skae274. J Anim Sci. 2024. PMID: 39279199
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
References
-
- Abdelaal A. M., Sohal I. S., Iyer S. G., Sudarshan K., Orellana E. A., Ozcan K. E., et al. (2024). Selective targeting of chemically modified miR-34a to prostate cancer using a small molecule ligand and an endosomal escape agent. Mol. Ther. Nucleic Acids 35 (2), 102193. 10.1016/j.omtn.2024.102193 - DOI - PMC - PubMed
-
- Amstalden V. H. E., Blackwell T. R., Klinkert I., Eijkel G. B., Heeren R. M., Glunde K. (2010). Multimodal mass spectrometric imaging of small molecules reveals distinct spatio-molecular signatures in differentially metastatic breast tumor models. Cancer Res. 70 (22), 9012–9021. 10.1158/0008-5472.CAN-10-0360 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

