Modelling the melting of DNA oligomers with non-inert dangling ends
- PMID: 40880986
- PMCID: PMC12380575
- DOI: 10.3389/fmolb.2025.1646428
Modelling the melting of DNA oligomers with non-inert dangling ends
Abstract
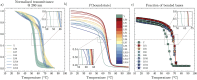
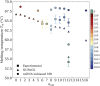
In this work, we investigate the dependence of the melting temperature of low-valency DNA constructs on the length of non-inert dangling ends, controlling their sequence composition. We compare two computational models to evaluate their effectiveness and limitations in predicting the melting behavior of DNA oligomers (bivalent linkers) and more complex structures (trivalent nanostars), benchmarking the results against experimental spectroscopic data. Our results suggest that the length of non-inert dangling ends has minimal impact on the melting point of the DNA duplex for the duplexes we studied, informing the future design of DNA supramolecular constructs.
Keywords: DNA nanotechnology; NUPACK; melting curves; molecular dynamics; oxDNA; umbrella sampling.
Copyright © 2025 Soto, Mambretti, Locatelli and Stoev.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

