Research progress on NAT10-mediated acetylation in normal development and disease
- PMID: 40881352
- PMCID: PMC12380694
- DOI: 10.3389/fcell.2025.1623276
Research progress on NAT10-mediated acetylation in normal development and disease
Abstract
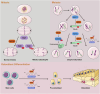
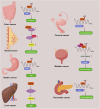
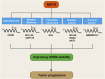
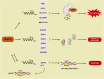
N4-acetylcytidine (ac4C) is an evolutionarily conserved RNA modification catalyzed by the acetyltransferase NAT10. It regulates RNA stability, translation, and post-transcriptional processes. Meanwhile, NAT10 functions as a dual-function enzyme exhibiting both protein acetyltransferase and RNA acetylase activities. This review summarizes the structural and functional roles of NAT10-mediated acetylation in physiological contexts, including cell division, differentiation, inflammation, aging, and viral infection, as well as its emerging roles in cancer. In malignancies, NAT10-mediated acetylation drives tumor progression by enhancing mRNA stability, regulating cell cycle, promoting metastasis, suppressing ferroptosis, modulating metabolism, influencing p53 activity, mediating immune escape and fostering drug resistance. Interactions between NAT10 and non-coding RNAs further amplify its oncogenic effects. Unresolved questions, such as microbiota-mediated ac4C regulation and NAT10's impact on the tumor immune microenvironment, highlight future research directions. Targeting NAT10 and ac4C modification presents promising therapeutic opportunities, with advanced technologies like single-cell sequencing poised to refine epitranscriptome-based interventions.
Keywords: AC4C; NAT10; RNA modification; cancer; epitranscriptomics; therapeutic target.
Copyright © 2025 Qin, Liu, Ma, Wang, Lu, Yang, Tang and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Emerging role of N-acetyltransferase 10 in diseases: RNA ac4C modification and beyond.Mol Biomed. 2025 Jul 1;6(1):46. doi: 10.1186/s43556-025-00286-3. Mol Biomed. 2025. PMID: 40588654 Free PMC article. Review.
-
Acetyltransferase NAT10 inhibits T-cell immunity and promotes nasopharyngeal carcinoma progression through DDX5/HMGB1 axis.J Immunother Cancer. 2025 Feb 12;13(2):e010301. doi: 10.1136/jitc-2024-010301. J Immunother Cancer. 2025. PMID: 39939141 Free PMC article.
-
NAT10 Mediates Cardiac Fibrosis Induced by Myocardial Infarction Through ac4C Modification of TGFBR1 mRNA.Cell Biol Toxicol. 2025 Aug 12;41(1):125. doi: 10.1007/s10565-025-10081-z. Cell Biol Toxicol. 2025. PMID: 40794120 Free PMC article.
-
Yin Yang 1 protein-activated N-acetyltransferase 10 drives cell malignant progression of osteosarcoma through ac4C acetylation of integrin β3.J Bone Oncol. 2025 Jul 12;53:100701. doi: 10.1016/j.jbo.2025.100701. eCollection 2025 Aug. J Bone Oncol. 2025. PMID: 40717796 Free PMC article.
-
The role and mechanism of NAT10-mediated ac4C modification in tumor development and progression.MedComm (2020). 2024 Dec 4;5(12):e70026. doi: 10.1002/mco2.70026. eCollection 2024 Dec. MedComm (2020). 2024. PMID: 39640362 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

