Combination of serology and PCR analysis of environmental samples to assess Coxiella burnetii infection status in small ruminant farms
- PMID: 40882006
- PMCID: PMC12542757
- DOI: 10.1128/aem.00931-25
Combination of serology and PCR analysis of environmental samples to assess Coxiella burnetii infection status in small ruminant farms
Abstract
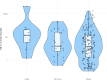
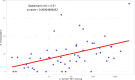
Coxiella burnetii, the causative agent of Q fever, accumulates on dust of farm premises with infected animals, but the interpretation of PCR detection on dust is challenging. To investigate whether bacterial load in environmental dust together with the within-flock seropositivity could be indicative of the C. burnetii infection status in small-ruminant flocks, 249 farms (202 sheep, 18 goats, and 29 sheep-goat mixed) in the Balearic Islands were investigated. Dust samples were analyzed by real-time PCR targeting C. burnetii IS1111, and C. burnetii loads (genome equivalents/mg dust) were estimated by quantitative real-time PCR amplification of com1 to categorize the farms into four levels. Sera from 546 yearlings and 1,002 adult females in 79 flocks were tested by ELISA. Despite a widespread distribution of C. burnetii in environmental dust, only 6.0% and 16.1% had high (>1,000) or moderate (100-1,000) C. burnetii loads, respectively. These same farms had significantly higher within-flock percentage of C. burnetii seropositive animals (~35%) than flocks with low loads or C. burnetii-negative. These results and the positive correlation observed between within-flock percentage of seropositive animals and C. burnetii environmental contamination indicate that dust PCR followed by serological analysis of flocks with high and moderate loads could be used to identify herds potentially infected with C. burnetii. SNP genotyping of Coxiella PCR-positive dust samples identified SNP-6 as the predominant genotype in small ruminants in the Balearic Islands, along with the sporadic presence of SNP-4-a clearly different genotype distribution than in northern Spain, where the clinical spectrum of human Q fever is clearly different.IMPORTANCEThe identification of flocks with active C. burnetii infection is crucial to implement control measures and prevent human Q fever cases. This study demonstrates the relevance of combining dust PCR with serology to identify C. burnetii-infected herds, a strategy that could help to identify the animal source of human Q fever outbreaks and define priority countermeasures. This study also provided, for the first time, an overview of C. burnetii infection in sheep and goats in the Balearic Islands and identified the factors associated with higher risk of environmental C. burnetii contamination. Strain characterization allowed the identification of the most prevalent C. burnetii genotypes in this region of eastern Spain, showing clear differences in genotype distribution with the northern area, which could explain the different clinical spectrum of human Q fever cases in both geographical areas.
Keywords: Coxiella burnetii; Q fever; environmental dust; quantitative PCR; real-time PCR; serology; small ruminants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
- Basque Governement/Department of Food, Rural Development, Agriculture and Fisheries
- BIA03/20-2/Fons de Garantia Agrària i Pesquera de les Illes Balears (FOGAIBA)
- PRE2018-087124/MCIN/AEI/10.13039/501100011033 and ESF Investing in your future
- Ramón y Cajal RYC2021-033084-I/MCIN/AEI and European Union NextGenerationEU/PRTR
LinkOut - more resources
Full Text Sources

