TNG260 Is a Small-Molecule CoREST Inhibitor That Sensitizes STK11-Mutant Tumors to Anti-PD-1 Immunotherapy
- PMID: 40882030
- PMCID: PMC12521918
- DOI: 10.1158/0008-5472.CAN-25-0998
TNG260 Is a Small-Molecule CoREST Inhibitor That Sensitizes STK11-Mutant Tumors to Anti-PD-1 Immunotherapy
Abstract
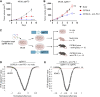
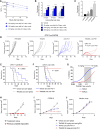
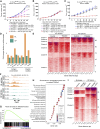
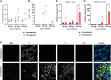
Patients with non-small cell lung cancer (NSCLC) with loss of the tumor suppressor gene STK11 are resistant to immune checkpoint therapies like anti-PD-1. In this study, we conducted an in vivo CRISPR screen that identified histone deacetylase 1 as a target to reverse anti-PD-1 resistance driven by loss of STK11 and developed TNG260, a potent small-molecule inhibitor of the CoREST complex with selectivity exceeding previously generated inhibitors in this class in preclinical studies. Treatment with TNG260 led to increased expression of immunomodulatory genes in STK11-deficient cancer cells. When combined with anti-PD-1, TNG260 induced immune-mediated stasis and/or regression in STK11-deficient syngeneic tumor models and autochthonous NSCLC models. In the tumors of patients with STK11-deficient cancers in a clinical trial (NCT05887492), treatment with a combination of TNG260 and pembrolizumab increased intratumoral histone acetylation, PD-L1 tumor proportion scores, and T-cell infiltration into the tumor microenvironment. This study illustrates a promising treatment strategy for addressing immune evasion in patients with STK11-mutant NSCLC.
Significance: Targeting CoREST with TNG260 sensitizes STK11-deficient non-small cell lung cancer to anti-PD-1 immunotherapy, offering a potential treatment for patients not served by existing therapies. See related commentary by Lin and Shen, p. 3821.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
L.G. Ahronian reports personal fees from Tango Therapeutics during the conduct of the study, as well as a patent for WO2024030659A1 issued to Tango Therapeutics. M. Zhang reports other support from Tango Therapeutics outside the submitted work. G.S. Falchook reports other support from Tango Therapeutics during the conduct of the study, as well as other support from Wolters Kluwer (royalties), other support from AbbVie (advisory role), Fujifilm (advisory role), Silicon (advisory role), Navire (advisory role), Turning Point (advisory role), Predicine (advisory role), Inspirna (advisory role), Regeneron (advisory role), Jubilant (advisory role), BostonGene (advisory role), Teon (advisory role), Merck (advisory role), Sanofi (advisory role), BridgeBio (advisory role), Avistone (advisory role), and EMD Serono (advisory role), other support from Total Health Conferencing (honorarium), Rocky Mountain Oncology Society (honorarium), and Clinical Care Options (honorarium), other support from Amgen (travel support), Bristol Myers Squibb (travel support), EMD Serono (travel support), Fujifilm (travel support), Millennium (travel support), Sarah Cannon Research Institute (travel support), Synthorx/Sanofi (travel support), GSK (travel support), and Cyteir (travel support), and other support from 3-V Biosciences, Abbisko, AbbVie, ABL Bio, ADC Therapeutics, Accutar, Agenus, Aileron, Alterome, American Society of Clinical Oncology, Amgen, Arcus, ARMO/Eli Lilly and Company, Artios, Astellas, AstraZeneca, Bayer, BeiGene, Beijing Avistone, Bioatla, Bioinvent, Biomea Fusion, Biothera, Bicycle, Black Diamond, Boehringer Ingelheim, Boundless, Celgene, Celldex, Centessa, Ciclomed, Conjupro, Curegenix, Curis, Cyteir, Cytomx, D3 Bio, Daiichi Sankyo, Deciphera, DelMar, Dynamicure, eFFECTOR, Eikon, Eli Lilly and Company, EMD Serono, Epizyme, Erasca, Exelixis, Freenome, Fujifilm, Genmab, GlaxoSmithKline, Harbour BioMed, Hutchison MediPharma, IGM Biosciences, IDEAYA, Ignyta, Ikena, Immuneering, Immunitas, ImmunoGen/MacroGenics, Incyte, Jacobio, Jazz, Jounce, Jubilant, Kineta, Kolltan, Kumquat, Kura, Loxo/Bayer, Medilink, MedImmune, Merck, Metabomed, Millennium, Mirati, miRNA Therapeutics, ModeX, Molecular Templates, Nammi, NIH, Navire/BridgeBio, NGM Bio, NiKang, Novartis, Nuvalent, Nuvectis, OncoMed, Oncusp, Oncorus, Oncothyreon, OnKure, Phanes, Poseida, Precision Oncology, Prelude, PureTech, Pyramid, Pyxis, Quanta, RasCal, Regeneron, Relay, Rgenix, Ribon, Roche, Samumed, Sapience, Sarah Cannon Development Innovations, Seagen, Silicon/Stingthera, Simcha, Sirnaomics, Strategia, Syndax, Synthorx/Sanofi, Taiho, Tachyon, Takeda, Tallac, Tango, Tarus, Tarveda, Teneobio, Tesaro, Tocagen, TORL, Turning Point, University of Texas MD Anderson Cancer Center, Vegenics, Xencor, and Zhuhai Yufan. J.W. Goldman reports grants from Tango Therapeutics during the conduct of the study, as well as grants and personal fees from AstraZeneca, AbbVie, and Eli Lilly and Company, personal fees from Genentech, and grants from Bristol Myers Squibb and Agenus outside the submitted work. A.I. Spira reports grants from Tango Therapeutics during the conduct of the study, as well as grants from Tango Therapeutics outside the submitted work. S.R. Punekar reports nonfinancial support from Tango Therapeutics during the conduct of the study, as well as nonfinancial support from Tango Therapeutics outside the submitted work. D.R. Spigel reports grants from Tango Therapeutics during the conduct of the study, as well as grants and other support from AbbVie, AstraZeneca, Roche/Genentech, GlaxoSmithKline, and Lyell Immunopharma, grants from Agios, Arcus, Ascendis Pharma, Asher Biotherapeutics, BeiGene, Beijing Avistone Biotechnology, Bicara Therapeutics, BioAtla, Blueprint Medicine, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Chugai, Cyteir Therapeutics, Ellipses Pharma, Erasca, Gilead Sciences, Janux Therapeutics, Jazz Pharmaceuticals, Kronos Bio, Kumquat Biosciences, Loxo Oncology, MacroGenics, Merck, Millennium Pharmaceuticals, Moderna, Molecular Template, Monte Rosa Therapeutics, NGM Biopharmaceuticals, Peloton Therapeutics, Phanes Therapeutics, ProfoundBio, PureTech Health, Razor Genomics, Repare Therapeutics, Rgenix, Scorpion Therapeutics, Shenzhen Chipscreen Biosciences, Stemline Therapeutics, Synthekine, Taiho, Tango Therapeutics, and Zai Laboratory, and other support from Amgen, Circle Pharma, Daiichi Sankyo, Gilead Sciences, MedImmune, ModeX Therapeutics, Ottimo Pharma, and Pyxis Oncology outside the submitted work. J.S. Wang reports other support from Tango Therapeutics during the conduct of the study, as well as other support from AbbVie, Abdera Therapeutics, Accent Therapeutics, Accutar Biotech, Acrivon Therapeutics, Adagene, Allorion Therapeutics, Alterome Therapeutics, Apollo, Artios, Astellas Pharma, BeiGene, Bicycle Therapeutics, BioNTech SE, Biostar, Blueprint Medicines, Bristol Myers Squibb GmbH & Co. KG, Boehringer Ingelheim, C4 Therapeutics, Celgene/Bristol Myers Squibb, Circle Pharma, Compass Therapeutics, Compugen, Cullinan Oncology, Conjupro Biotherapeutics, D3 Bio, Daiichi Sankyo/UCB Japan, Day One Bio, Dren Bio, DualityBio, Edgewood Oncology, Ellipses Pharma, Erasca, Inc, Genentech/Roche, Genmab, Georgiamune, GlaxoSmithKline, Halda Therapeutics, Hotspot Therapeutics, IgM Biosciences, Immunitas, Immunogen, Incyte, ITeos Therapeutics, Janssen, Jazz Pharmaceuticals, Kineta, Klus Pharma, Kumquat, Kura Oncology, Loxo/Lilly, MabSpace Biosciences, Macrogenics, MBQ Pharma, Medikine, MediLink Therapeutics, Stemline/Menarini, Merck KGaA, Mersana, Moderna Therapeutics, NGM Biopharmaceuticals, NiKang, Novartis, Nurix, Olema Oncology, OnCusp Therapeutics, Pfizer, Pyxis, Quanta Therapeutics, Relay Therapeutics, Revolution Medicines, Sanofi, Step Pharma, Syndax, Systimmune, Vividion Therapeutics, Xencor, Zai Lab, and Zymeworks. F. Skoulidis reports grants, personal fees, and other support from Amgen, Revolution Medicines, and Bristol Myers Squibb, grants and personal fees from Merck & Co and Novartis, personal fees from BridgeBio, BeiGene, BergenBio, Guardant Health, Calithera Biosciences, Tango Therapeutics, Roche, Novocure, Hookipa Pharma, Regeneron, ESMO, Japanese Lung Cancer Society, Medscape LLC, Intellisphere LLC, personal fees and other support from AstraZeneca, AACR, IASLC, MJH Life Sciences, IDEOlogy Health, MI&T, PER LLC, and CURIO LLC, and other support from DAVA Oncology outside the submitted work. Y. Yu reports personal fees from Tango Therapeutics during the conduct of the study, as well as ownership of Tango Therapeutics stocks. P. McCarren reports other support from Tango Therapeutics during the conduct of the study. A. Tsai reports personal fees from Tango Therapeutics during the conduct of the study. P. Shahagadkar reports personal fees from Tango Therapeutics during the conduct of the study. N.M. Das reports personal fees from Tango Therapeutics during the conduct of the study. L. Danielczyk reports other support from Tango Therapeutics outside the submitted work. S.R. Meier reports other support from Tango Therapeutics outside the submitted work. D.A. Whittington reports personal fees and other support from Sesame Therapeutics outside the submitted work. C. Min reports other support from Tango Therapeutics outside the submitted work, as well as a patent for WO2024030659A1 pending. I. Sienczylo reports personal fees from Tango Therapeutics during the conduct of the study, as well as a patent 631137 pending. J.P. Maxwell reports personal fees and nonfinancial support from Tango Therapeutics and nonfinancial support from Sesame Therapeutics outside the submitted work, as well as a patent for US 12043607 issued. H.J. DiBenedetto reports other support from Tango Therapeutics during the conduct of the study. A. Crystal reports personal fees and other support from Tango Therapeutics during the conduct of the study, as well as personal fees and other support from Tango Therapeutics outside the submitted work. J.N. Andersen reports personal fees and other support from Tango therapeutics outside the submitted work. K.-K. Wong reports grants from Tango Therapeutics during the conduct of the study, as well as grants from Janssen Pharmaceuticals, Pfizer, Bristol Myers Squibb, Zentalis Pharmaceuticals, Blueprint Pharmaceuticals, Takeda, Novartis, Genentech, BridgeBio Pharma, Boehringer Ingelheim, Cogent, Revolution Medicine, and AstraZeneca and personal fees from Allerion Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Figures

References
-
- Alexander B, Sokol ES, Danziger NA, Pavlick DC, Elvin J, Killian JK, et al. 107P immune checkpoint inhibitor (ICPI) resistance genes STK11 and KEAP1: a comparative comprehensive genomic profiling (CGP) study. Ann Oncol 2020;31:S283–4.
-
- Heist RS, Yu J, Donderici EY, Zhang N, Espenschied CR, Lang K, et al. Impact of STK11 mutation on first-line immune checkpoint inhibitor (ICI) outcomes in a real-world KRAS G12C mutant lung adenocarcinoma cohort. J Clin Oncol 2021;39:9106.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

