An anti-CEA affibody showing high-definition staining in human pancreatic cancer tissue sections and selective tumor targeting in vivo
- PMID: 40882560
- PMCID: PMC12410180
- DOI: 10.1016/j.tranon.2025.102512
An anti-CEA affibody showing high-definition staining in human pancreatic cancer tissue sections and selective tumor targeting in vivo
Abstract
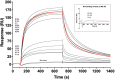
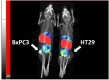
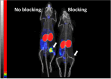
We report development and characterization of small non-immunoglobulin affibody affinity proteins directed to the highly glycosylated human carcinoembryonic antigen-related adhesion molecule 5 (CEACAM5, CEA), and their use in immunohistochemical (IHC) analyses of human pancreatic cancer samples and for in vivo tumor imaging. A total of nineteen unique anti-CEA affibodies were identified from large phage display libraries constructed using combinatorial protein engineering of a small 58 amino acid three-helix bundle protein domain. Molecular modeling suggested that all enriched clones share a binding surface with several clustered tryptophan residues interacting with a hydrophobic patch in the N1 domain of CEA centered around a phenylalanine residue. One variant, designated as C9, exhibited the highest affinity in biosensor analyses and was reformatted into a 15 kDa homodimer expressed in Escherichia coli. The biotinylated form, C9-C9-Bio, was evaluated for its IHC performance on matched frozen and formalin-fixed, paraffin-embedded (FFPE) sections of human pancreatic cancer samples (n = 7). Compared to clinical-grade monoclonal antibodies II-7 and CEA31, as well as a polyclonal reagent, C9-C9-Bio demonstrated highly sensitive CEA detection with minimal background staining. Statistical analyses including intraclass correlation and Bland-Altman assessments revealed excellent agreement between C9-C9-Bio and the two monoclonal antibodies in FFPE tissue samples. Further, a 99mTc[Tc]-labeled C9-C9 construct showed CEA-dependent binding to human cancer cell lines in vitro, and selectively bound to CEA-expressing BxPC3 xenografts in mice when investigated as a tracer for in vivo imaging, allowing for a visualization of tumors after four hours. In summary, these findings highlight the potential use of the easily produced CEA-binding C9 affibody for various clinical applications, including IHC and medical imaging, and as a targeting moiety for directing various therapeutic modalities to CEA-expressing tumors.
Keywords: Adenocarcinoma; Affibody; Affinity; CEA; CEA31; CEACAM5; Carcinoembryonic antigen; Clinical diagnostics; HRP; IHC; II-7; Immunohistochemistry; In vivo imaging; Pancreatic cancer; Phage display; Xenograft.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The new anti-CEA affinity proteins described in the manuscript are subject of a patent application, involving the following inventors: JN, CFM, HS, AA, MB and PÅN.
Figures






References
-
- Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A., Ronneberger O., Willmore L., Ballard A.J., Bambrick J., Bodenstein S.W., Evans D.A., Hung C.C., O'Neill M., Reiman D., Tunyasuvunakool K., Wu Z., Zemgulyte A., Arvaniti E., Beattie C., Bertolli O., Bridgland A., Cherepanov A., Congreve M., Cowen-Rivers A.I., Cowie A., Figurnov M., Fuchs F.B., Gladman H., Jain R., Khan Y.A., Low C.M.R., Perlin K., Potapenko A., Savy P., Singh S., Stecula A., Thillaisundaram A., Tong C., Yakneen S., Zhong E.D., Zielinski M., Zidek A., Bapst V., Kohli P., Jaderberg M., Hassabis D., Jumper J.M. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024 - PMC - PubMed
-
- Ahlgren S., Andersson K., Tolmachev V. Kit formulation for 99mTc-labeling of recombinant anti-HER2 affibody molecules with a C-terminally engineered cysteine. Nucl. Med. Biol. 2010;37:539–546. - PubMed
-
- Altena R., Buren S.A., Blomgren A., Karlsson E., Tzortzakakis A., Brun N., Moein M.M., Jussing E., Frejd F.Y., Bergh J., Tran T.A., Hartman J., Axelsson R. Human epidermal growth factor receptor 2 (HER2) PET imaging of HER2-low breast cancer with [(68)Ga]Ga-ABY-025: results from a pilot study. J. Nucl. Med. 2024;65:700–707. - PubMed
-
- Baker M. Reproducibility crisis: blame it on the antibodies. Nature. 2015;521:274–276. - PubMed
-
- Ballehaninna U.K., Chamberlain R.S. Biomarkers for pancreatic cancer: promising new markers and options beyond CA 19-9. Tumour. Biol. 2013;34:3279–3292. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

