Gene Expression Insights into Cytarabine Resistance in Acute Myeloid Leukemia: The Role of Cytokines
- PMID: 40883022
- PMCID: PMC12402712
- DOI: 10.21873/cgp.20533
Gene Expression Insights into Cytarabine Resistance in Acute Myeloid Leukemia: The Role of Cytokines
Abstract
Background/aim: Cytarabine is the main chemotherapy agent used to treat acute myeloid leukemia (AML), but drug resistance remains a major challenge. Imbalances in cytokine secretion are known to play a role in the survival and proliferation of AML blast cells. While numerous studies have investigated cytokine secretion in AML, the precise role of cytokines in the pathogenesis of AML remains unclear. This study aimed to compare cytokine-related gene expression in parental and resistant HL60 cells and assess their prognostic relevance in AML.
Materials and methods: This study compared gene expression profiles of parental HL60 and R-HL60 cells by analyzing gene expression arrays. Further, the correlation between the differential expression genes (DEGs) and overall survival in patients with AML was obtained from the Cancer Genome Atlas (TCGA) database, and the expression of genes was validated by quantitative PCR (QPCR).
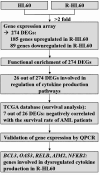
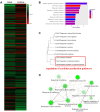
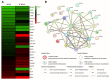
Results: Our results showed that twenty-six genes involved in cytokine regulation were significantly different between HL60 and R-HL60 cells. The DEGs associated with cytokine production between parental HL60 and R-HL60 was subjected to a functional enrichment analysis by ShinyGO. The expression of BCL3, OAS3, RELB, AIM2, NFKB2, CLEC9A, and OAS2 involved in the regulation of cytokine production was associated with survival probability of AML patient in the TCGA database. Among them, the expression of BCL3, OAS3, RELB, AIM2, and NFKB2 genes in R-HL60 cells was higher than that in HL-60 cells.
Conclusion: This study identified key genes involved in cytokine dysregulation in cytarabine-resistant HL60 cells, providing potential targets for overcoming drug resistance in AML. These findings offer new avenues for the development of more effective therapies for relapsed or refractory AML patients.
Keywords: Acute myeloid leukemia; HL60; cytarabine-resistance; cytokine production.
Copyright © 2025, International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Conflict of interest statement
The Authors have no conflicts of interest to declare.
Figures


Similar articles
-
Clinical Characteristics and Outcomes of Acute Myeloid Leukemia Patients Harboring NPM1/FLT3-ITD/DNMT3A Triple Mutations and the Potential Prognostic Value of GNG4.Cancer Control. 2025 Jan-Dec;32:10732748251359836. doi: 10.1177/10732748251359836. Epub 2025 Jul 17. Cancer Control. 2025. PMID: 40676766 Free PMC article.
-
Cytarabine Pharmacogenomics and Outcomes Among Children and Young Adults With Acute Myeloid Leukemia.JAMA Netw Open. 2025 Jun 2;8(6):e2516296. doi: 10.1001/jamanetworkopen.2025.16296. JAMA Netw Open. 2025. PMID: 40549387 Free PMC article. Clinical Trial.
-
Unveiling the role of coagulation-related genes in acute myeloid leukemia prognosis and immune microenvironment through machine learning.Eur J Med Res. 2025 Aug 11;30(1):734. doi: 10.1186/s40001-025-02975-9. Eur J Med Res. 2025. PMID: 40790216 Free PMC article.
-
Efficacy of non-intensive therapies approved for relapsed/refractory acute myeloid leukemia: a systematic literature review.Future Oncol. 2022 May;18(16):2029-2039. doi: 10.2217/fon-2021-1355. Epub 2022 Feb 24. Future Oncol. 2022. PMID: 35196866
-
Therapies for acute myeloid leukemia in patients ineligible for standard induction chemotherapy: a systematic review.Future Oncol. 2023 Apr;19(11):789-810. doi: 10.2217/fon-2022-1286. Epub 2023 May 12. Future Oncol. 2023. PMID: 37170899
References
-
- Laszlo GS, Ries RE, Gudgeon CJ, Harrington KH, Alonzo TA, Gerbing RB, Raimondi SC, Hirsch BA, Gamis AS, Meshinchi S, Walter RB. High expression of suppressor of cytokine signaling-2 predicts poor outcome in pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. Leuk Lymphoma. 2014;55(12):2817–2821. doi: 10.3109/10428194.2014.893305. - DOI - PMC - PubMed
-
- Sanchez-Correa B, Bergua JM, Campos C, Gayoso I, Arcos MJ, Bañas H, Morgado S, Casado JG, Solana R, Tarazona R. Cytokine profiles in acute myeloid leukemia patients at diagnosis: Survival is inversely correlated with IL-6 and directly correlated with IL-10 levels. Cytokine. 2013;61(3):885–891. doi: 10.1016/j.cyto.2012.12.023. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
