Altered development and network connectivity in a human neuronal model of 15q11.2 deletion-related neurodevelopmental disorders
- PMID: 40883268
- PMCID: PMC12397418
- DOI: 10.1038/s41398-025-03453-w
Altered development and network connectivity in a human neuronal model of 15q11.2 deletion-related neurodevelopmental disorders
Abstract
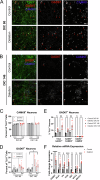
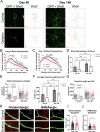
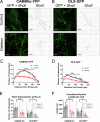
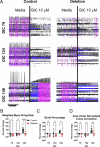
The chromosome 15q11.2 locus is deleted in 1.5% of patients with genetic epilepsy and confers a risk for intellectual disability and schizophrenia. Individuals with this deletion demonstrate increased cortical thickness, decreased cortical surface area and white matter abnormalities. Human induced pluripotent stem cell (iPSC)-derived neural progenitor cells from 15q11.2 deletion individuals exhibit early adhesion junction and migration abnormalities, but later neuronal development and function have not been fully assessed. Imaging studies indicating altered structure and network connectivity in the anterior brain regions and the cingulum suggest that in addition to alterations in neural progenitor dynamics, there may also be structural and functional changes within discrete networks of neurons. To explore this, we generated human forebrain cortical neurons from iPSCs derived from individuals with or without 15q11.2 deletion and used longitudinal imaging and multielectrode array analysis to evaluate neuronal development over time. 15q11.2 deleted neurons exhibited fewer connections and an increase in inhibitory neurons. Individual neurons had decreased neurite complexity and overall decreased neurite length. These structural changes were associated with a reduction in multiunit action potential generation, bursting and synchronization. The 15q11.2 deleted neurons also demonstrated specific functional deficits in glutamate- and GABA-mediated neuronal network activity and synchronization with a delay in the maturation of the inhibitory response to GABA. These data indicate that deletion of the 15q11.2 region is sufficient to impair the structural and functional maturation of cortical neuron networks, and suggest an in vitro correlate to the pathologic changes in humans with the 15q11.2 deletion.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics and consent to participate: De-identified fibroblast samples and iPSCs were obtained from commercial sources. Informed consent for sample donation was obtained at the Coriell Institute Biorepositories (Camden, New Jersey) and Cedar Sinai (West Hollywood, California) prior to deidentification and material transfer. All methods were performed in accordance with institutional safety and internal review board guidelines.
Figures


Update of
-
Altered development and network connectivity in a human neuronal model of 15q11.2 deletion-related neurodevelopmental disorders.bioRxiv [Preprint]. 2024 Sep 20:2024.09.19.613912. doi: 10.1101/2024.09.19.613912. bioRxiv. 2024. Update in: Transl Psychiatry. 2025 Aug 29;15(1):329. doi: 10.1038/s41398-025-03453-w. PMID: 39345567 Free PMC article. Updated. Preprint.
References
-
- Caseras X, Kirov G, Kendall KM, Rees E, Legge SE, Bracher-Smith M, et al. Effects of genomic copy number variants penetrant for schizophrenia on cortical thickness and surface area in healthy individuals: analysis of the UK biobank. Br J Psychiatry. 2021;218:104–11. 10.1192/bjp.2020.139 - PMC - PubMed
MeSH terms
Grants and funding
- R35 NS097370/NS/NINDS NIH HHS/United States
- K08NS102526/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 NS117604/NS/NINDS NIH HHS/United States
- K08 NS102526/NS/NINDS NIH HHS/United States
- R35 NS137480/NS/NINDS NIH HHS/United States
- R35NS116843/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- Clinical Scientist Award/Doris Duke Charitable Foundation (DDCF)
- R35NS097370/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- W81XWH1810175/U.S. Department of Defense (United States Department of Defense)
- CNCDPK12/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R25N065729/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R35 NS116843/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources

