Prefoldin complex promotes interferon-stimulated gene expression and is inhibited by rotavirus VP3
- PMID: 40883306
- PMCID: PMC12397275
- DOI: 10.1038/s41467-025-63393-3
Prefoldin complex promotes interferon-stimulated gene expression and is inhibited by rotavirus VP3
Abstract
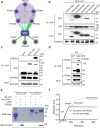
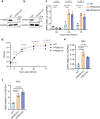
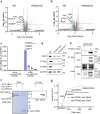
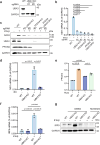
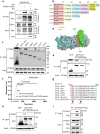
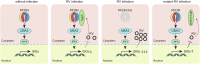
Timely induction of interferons and interferon-stimulated genes (ISGs) is critical for successful host defense against viral infections. VP3 encoded by rotavirus is implicated in interferon antagonism. However, the precise mechanisms remain incompletely understood. By conducting tandem-affinity purification coupled with high-resolution mass spectrometry, we identify the prefoldin complex as the top cellular binding partner of VP3. Rotavirus infection is significantly enhanced in prefoldin subunit knockout cells. Using proteome-wide label-free quantification, we find that prefoldin assists in folding ubiquitin-like-modifier-activating-enzyme-3 (UBA3), both of which positively regulate ISG expression. Through direct and competitive binding, VP3 interferes with the chaperone activity of prefoldin, leading to unstable UBA3, reduces IRF9, and suppresses ISG transcription. Our findings report a novel function of a prefoldin-UBA3-IRF9-ISG axis in antiviral immunity and uncover new aspects of virus-host interactions that could be exploited for broad-spectrum antiviral therapeutic development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Simons, C. T. et al. Selective contribution of eukaryotic prefoldin subunits to actin and tubulin binding. J. Biol. Chem.279, 4196–4203 (2004). - PubMed
-
- Balchin, D., Milicic, G., Strauss, M., Hayer-Hartl, M. & Hartl, F. U. Pathway of Actin Folding Directed by the Eukaryotic Chaperonin TRiC. Cell174, 1507–1521.e1516 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

