A whole-brain male mouse atlas of long-range inputs to histaminergic neurons
- PMID: 40883329
- PMCID: PMC12397332
- DOI: 10.1038/s41467-025-63394-2
A whole-brain male mouse atlas of long-range inputs to histaminergic neurons
Abstract
The precise structural and functional characteristics of input circuits targeting histaminergic neurons remain poorly understood. Here, using a rabies virus retrograde tracing system combined with fluorescence micro-optical sectioning tomography, we construct a 3D monosynaptic long-range input atlas of male mouse histaminergic neurons. We identify that the hypothalamus, thalamus, pallidum, and hippocampus constitute major input sources, exhibiting diverse spatial distribution patterns and neuronal type ratios. Notably, a specific layer distribution pattern and co-projection structures of upstream cortical neurons are well reconstructed at single-cell resolution. As histaminergic system is classically involved in sleep-wake regulation, we demonstrate that the lateral septum (predominantly supplying inhibitory inputs) and the paraventricular nucleus of the thalamus (predominantly supplying excitatory inputs) establish monosynaptic connections, exhibiting distinct functional dynamics and regulatory roles in rapid-eye-movement sleep. Collectively, our study provides a precise long-range input map of mouse histaminergic neurons at mesoscopic scale, laying a solid foundation for future systematic study of histaminergic neural circuits.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

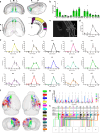
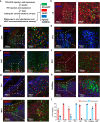

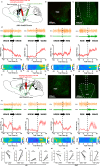

References
-
- Yoshikawa, T., Nakamura, T. & Yanai, K. Histaminergic neurons in the tuberomammillary nucleus as a control centre for wakefulness. Br. J. Pharmacol. 10.1111/bph.15220 (2020). - PubMed
-
- de Almeida, M. A. & Izquierdo, I. Memory facilitation by histamine. Arch. Int. Pharmacodyn. Ther.283, 193–198 (1986). - PubMed
MeSH terms
Substances
Grants and funding
- U21A20418/National Natural Science Foundation of China (National Science Foundation of China)
- 82300818/National Natural Science Foundation of China (National Science Foundation of China)
- U23A20533/National Natural Science Foundation of China (National Science Foundation of China)
- T2122015/National Natural Science Foundation of China (National Science Foundation of China)
- 2023M733191/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Research Materials

