Liver fat quantification at 0.55 T enabled by locally low-rank enforced deep learning reconstruction
- PMID: 40883956
- PMCID: PMC12620156
- DOI: 10.1002/mrm.70057
Liver fat quantification at 0.55 T enabled by locally low-rank enforced deep learning reconstruction
Abstract
Purpose: The emergence of new medications for fatty liver conditions has increased the need for reliable and widely available assessment of MRI proton density fat fraction (MRI-PDFF). Whereas low-field MRI presents a promising solution, its utilization is challenging due to the low SNR. This work aims to enhance SNR and enable precise PDFF quantification at low-field MRI using a novel locally low-rank deep learning-based (LLR-DL) reconstruction.
Methods: LLR-DL alternates between regularized SENSE and a neural network (U-Net) throughout several iterations, operating on complex-valued data. The network processes the spectral projection onto singular value bases, which are computed on local patches across the echoes dimension. The output of the network is recast into the basis of the original echoes and used as a prior for the following iteration. The final echoes are processed by a multi-echo Dixon algorithm. Two different protocols were proposed for imaging at 0.55 T. An iron-and-fat phantom and 10 volunteers were scanned on both 0.55 and 1.5 T systems. Linear regression, t-statistics, and Bland-Altman analyses were conducted.
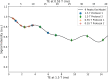
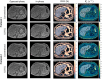
Results: LLR-DL achieved significantly improved image quality compared to the conventional reconstruction technique, with a 32.7% increase in peak SNR and a 25% improvement in structural similarity index. PDFF repeatability was 2.33% in phantoms (0% to 100%) and 0.79% in vivo (3% to 18%), with narrow cross-field strength limits of agreement below 1.67% in phantoms and 1.75% in vivo.
Conclusion: An LLR-DL reconstruction was developed and investigated to enable precise PDFF quantification at 0.55 T and improve consistency with 1.5 T results.
Keywords: deep learning; liver PDFF; locally low‐rank; low‐field MRI; reconstruction.
© 2025 The Author(s). Magnetic Resonance in Medicine published by Wiley Periodicals LLC on behalf of International Society for Magnetic Resonance in Medicine.
Conflict of interest statement
Majd Helo receives PhD funding from Siemens Healthineers AG. Dominik Nickel and Stephan Kannengiesser are employed by Siemens Healthineers AG.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

