G-quadruplex-dependent transcriptional regulation by molecular condensation in the Bcl3 promoter
- PMID: 40884402
- PMCID: PMC12397910
- DOI: 10.1093/nar/gkaf827
G-quadruplex-dependent transcriptional regulation by molecular condensation in the Bcl3 promoter
Abstract
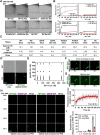
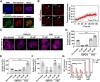
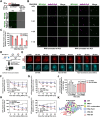
G-quadruplexes (G4s) are pivotal in transcriptional regulation. Although the interaction between G4s and G4-binding transcription factors (TFs) is critical for G4-dependent transcriptional regulation, the detailed mechanism, especially TF enrichment at G4s and its correlation with transcriptional regulation, remains unknown. In this study, using specificity protein 1 (SP1) as a representative G4-binding TF, we examined the mechanism of G4-dependent transcriptional regulation. Genomic analysis revealed substantial enrichment of SP1 in the oncogenic Bcl3 promoter harboring G4-forming sequences. We demonstrated that the formation of transcriptional condensates and the transcriptional activation of the Bcl3 promoter are heavily dependent on G4-dependent SP1 binding. Moreover, dissociation of SP1 condensates was prompted by RNA, which was enhanced by G4 formation within the RNA. Collectively, these results underscore the pivotal role of G4 in regulating gene expression through the modulation of SP1-mediated transcriptional condensation.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

