High-throughput competitive binding assay for targeting RNA tertiary structures with small molecules: application to pseudoknots and G-quadruplexes
- PMID: 40884404
- PMCID: PMC12397912
- DOI: 10.1093/nar/gkaf819
High-throughput competitive binding assay for targeting RNA tertiary structures with small molecules: application to pseudoknots and G-quadruplexes
Abstract
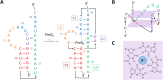
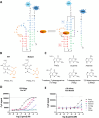
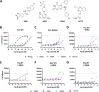
There is an indisputable need for new screening methodologies to identify small molecules that target RNA tertiary structures, such as pseudoknots or G-quadruplexes. Here, we present a high-throughput competitive binding antisense assay designed to identify ligands for complex RNA tertiary structures. In this assay, initially customized for the bacterial PreQ1-I riboswitch pseudoknot, ligands compete with a rationally designed quencher-labelled antisense oligonucleotide for binding to a fluorophore-labelled riboswitch. The method is validated for four PreQ1-I riboswitches, using the natural riboswitch ligand PreQ1 and various analogues. A commercial RNA-focused library consisting of ∼15 000 compounds was then screened against the Fusobacterium nucleatum riboswitch, leading to the identification of a promising hit, 4494, which showed competitive binding activity to all PreQ1-I riboswitches and was able to inhibit translation of a riboswitch-regulated reporter gene. Although resynthesis of 4494 revealed that its activity originated from an ∼1% guanine contamination, this result underscores the assay's exceptional sensitivity. To demonstrate its versatility, the assay was tailored for a SARS-CoV-2 G-quadruplex structure and validated with several known G-quadruplex ligands. This work shows that the competitive binding antisense assay is a powerful addition to the RNA-targeting toolbox, facilitating the discovery of ligands for diverse RNA tertiary structures.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures





Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Opportunities for Riboswitch Inhibition by Targeting Co-Transcriptional RNA Folding Events.Int J Mol Sci. 2024 Sep 29;25(19):10495. doi: 10.3390/ijms251910495. Int J Mol Sci. 2024. PMID: 39408823 Free PMC article. Review.
-
Tiny but multi-stable: four distinct conformational states govern the ligand-free state of the preQ1 riboswitch from a thermophilic bacterium.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf560. doi: 10.1093/nar/gkaf560. Nucleic Acids Res. 2025. PMID: 40586310 Free PMC article.
-
Two riboswitch classes that share a common ligand-binding fold show major differences in the ability to accommodate mutations.Nucleic Acids Res. 2024 Nov 27;52(21):13152-13173. doi: 10.1093/nar/gkae886. Nucleic Acids Res. 2024. PMID: 39413212 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

