Pathogenic variants in TMEM184B cause a neurodevelopmental syndrome associated with alteration of metabolic signaling
- PMID: 40885185
- PMCID: PMC12440709
- DOI: 10.1016/j.ajhg.2025.08.004
Pathogenic variants in TMEM184B cause a neurodevelopmental syndrome associated with alteration of metabolic signaling
Abstract
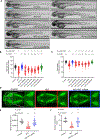
Transmembrane protein 184B (TMEM184B) is an endosomal 7-pass transmembrane protein with evolutionarily conserved roles in synaptic structure and axon degeneration. We report six pediatric cases who have de novo heterozygous variants in TMEM184B; five individuals harbor a rare missense variant, and one individual has an mRNA splice site change. This cohort is unified by overlapping neurodevelopmental deficits including developmental delay, corpus callosum hypoplasia, seizures, and/or microcephaly. TMEM184B is predicted to contain a pore domain wherein four of five human disease-associated missense variants cluster. Structural modeling suggests that all missense variants alter TMEM184B protein stability. To understand the contribution of TMEM184B to neural development in vivo, we knocked down the TMEM184B ortholog in zebrafish and observed microcephaly and reduced anterior commissural axons, aligning with symptoms of affected individuals. Ectopic expression of TMEM184B c.550A>G (p.Lys184Glu) and c.484G>A (p.Gly162Arg) variants cause reduced head size and body length, indicating dominant effects, while three other variants show haploinsufficiency. None of the variants are able to rescue the knockdown phenotype. Human induced pluripotent stem cells with monoallelic production of p.Lys184Glu show mRNA disruptions in key metabolic pathways including those controlling mechanistic target of rapamycin activity. Expression of p.Lys184Glu and c.863G>C (p.Gly288Ala) increased apoptosis in cell lines, and p.Lys184Glu increased nuclear localization of transcription factor EB, consistent with a cellular starvation state. Together, our data indicate that TMEM184B variants cause cellular metabolic disruption and result in abnormal neural development.
Keywords: TFEB; corpus callosum; dominant toxic; epilepsy; haploinsufficiency; lysosome; microcephaly; neurodevelopment; transmembrane protein; zebrafish.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.V.K. and S.S. are employed by AIQure LLC. S.S. is employed by Expert Systems Inc.
Figures



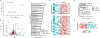


Update of
-
Pathogenic variants in TMEM184B cause a neurodevelopmental syndrome associated with alteration of metabolic signaling.medRxiv [Preprint]. 2025 Aug 22:2024.06.27.24309417. doi: 10.1101/2024.06.27.24309417. medRxiv. 2025. Update in: Am J Hum Genet. 2025 Oct 2;112(10):2381-2401. doi: 10.1016/j.ajhg.2025.08.004. PMID: 39006436 Free PMC article. Updated. Preprint.
References
-
- Cho TS, Beigaitė E, Klein NE, Sweeney ST, and Bhattacharya MRC (2022). The Putative Drosophila TMEM184B Ortholog Tmep Ensures Proper Locomotion by Restraining Ectopic Firing at the Neuromuscular Junction. Mol. Neurobiol. 10.1007/S12035-022-02760-3. - DOI
-
- Oliva CA, Montecinos-Oliva C, and Inestrosa NC (2018). Chapter Three - Wnt Signaling in the Central Nervous System: New Insights in Health and Disease. In Progress in Molecular Biology and Translational Science WNT Signaling in Health and Disease, Larraín J and Olivares G, eds. (Academic Press; ), pp. 81–130. 10.1016/bs.pmbts.2017.11.018. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

