Evidence that mitochondria in macrophages are destroyed by microautophagy
- PMID: 40885798
- PMCID: PMC12398504
- DOI: 10.1038/s41467-025-63531-x
Evidence that mitochondria in macrophages are destroyed by microautophagy
Abstract
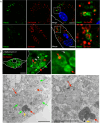
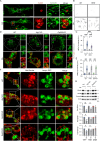
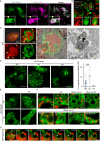
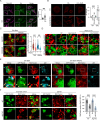
Microautophagy is an intracellular degradation process in which degradatory organelles, such as the lysosome, directly take up substrates by invagination and/or protrusion of their membranes. Here, we provide evidence that Rab32-positive, lysosome-related organelles in macrophages incorporate various other organelles, including endosomes and mitochondria. Our data indicates that, upon exposure to a mitochondria-damaging reagent, mitochondria can be directly engulfed by the lysosome-like organelles independently of macroautophagy or ESCRT machinery. Rab32 GTPase, phosphatidylinositol 3,5-bisphosphates, ubiquitination, and p62/SQSTM1 are crucial for this degradation. Furthermore, the degree of M1 polarization of macrophages, which is facilitated by metabolic reprogramming into increased glycolysis via mitochondrial elimination, is significantly reduced in Rab32/38 double-knockout macrophages. Thus, microautophagy plays a role in the physiological regulation of macrophages.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- Oku, M. & Sakai, Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays40, e1800008 (2018). - PubMed
-
- Mijaljica, D., Prescott, M. & Devenish, R. J. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy7, 673–682 (2011). - PubMed
MeSH terms
Substances
Grants and funding
- 20H05326/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22H04647/MEXT | Japan Society for the Promotion of Science (JSPS)
- 23H02475/MEXT | Japan Society for the Promotion of Science (JSPS)
- 20K15789/MEXT | Japan Society for the Promotion of Science (JSPS)
- 25K09629/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources

