Selective disruption of DNMT1/ELK1 interactions induces DGKI re-expression and promotes temozolomide sensitivity of MGMTmethylated/DGKImethylated glioblastoma
- PMID: 40886022
- PMCID: PMC12398979
- DOI: 10.1186/s13148-025-01943-8
Selective disruption of DNMT1/ELK1 interactions induces DGKI re-expression and promotes temozolomide sensitivity of MGMTmethylated/DGKImethylated glioblastoma
Abstract
Background: DNA methyltransferase (DNMT) inhibitors are emerging as a promising class of agents for personalized and targeted cancer therapy, particularly in malignancies with limited therapeutic options such as glioblastoma (GB). In GB, the MGMT/DGKI methylation profile serves as a biomarker for stratifying patients by treatment response. Specifically, the MGMTmethylated/DGKIunmethylated profile is associated with favorable outcomes, whereas the MGMTmethylated/DGKImethylated profile correlates with poor outcome. These findings suggest that selective demethylation of DGKI without altering MGMT or inducing widespread genomic hypomethylation, which may promote tumorigenesis, could represent a novel and more effective therapeutic strategy.
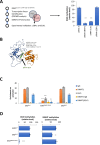
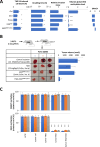
Results: Current DNMT inhibitors lack specificity for glioblastoma relevant methylation profiles, thereby limiting their therapeutic efficacy. To address this challenge, AlphaFold-based protein-protein interaction predictions were integrated with sequential chromatin immunoprecipitation assays and established DNMT1 interactome data. This integrative approach led to the identification of a DNMT1/ELK1 complex as a critical regulator of DGKI methylation. A peptide mimicking the DNMT1/ELK1 interface, designated EXDDNMT1/ELK1, was designed and shown to induce selective DGKI demethylation without altering MGMT or inducing global DNA hypomethylation. Notably, EXDDNMT1/ELK1 did note promote cellular proliferation or invasion, and successfully restored sensitivity to standard glioblastoma therapy in both cellular and in vivo models. These findings also support the use of MGMT and DGKI methylation levels in cell-free DNA as potential biomarkers to identify patients likely to benefit from EXDDNMT1/ELK1 treatment.
Conclusion: This study identifies a clinically actionable biomarker (MGMTMethylated/DGKIMethylated), detectable in both solid and liquid biopsies, enabling patient stratification. Furthermore, it establishes EXDDNMT1/ELK1 as a highly selective epigenetic therapeutic agent to treat GB patients.
Keywords: DNMT1; ELK1; Epidrug; Peptide; Protein–protein interaction.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Tumors were collected from the “French Glioblastoma Biobank FGB” [8]. The FGB network was declared to the French Ministry of Health and Research (declaration number: DC-2011-1467, cession authorization number: AC-2017-2993). The protocols and regulations of the FGB network were approved by the CPP OUEST II ethics committee (CB 2012/02, date of approval: 20 December 2011) and the CNIL (no. 1476342, date of approval: 10 October 2011). Plasma was collected from patients treated at the “Institut de Cancérologie de l’Ouest” (ICO, http://www.ico-cancer.fr ). All patients recruited gave signed and informed consent. All the samples collected and the associated clinical information were registered in the database (N° DC-2018-3321) validated by the French research ministry. Biological resources were stored at the “Centre de Ressources Biologiques-Tumorothèque (CRB)” (Institut de Cancérologie de l’Ouest, Saint-Herblain, F44800, France) [9]. Consent for publication: All authors have given consent for publication. Competing interests: The authors declare no competing interests.
Figures


References
-
- Tamimi AF, Juweid M. Epidemiology and Outcome of Glioblastoma. In: De Vleeschouwer S, editor. Glioblastoma [Internet]. Brisbane (AU): Codon Publications. 2017. Available from: http://www.ncbi.nlm.nih.gov/books/NBK470003/. - PubMed
-
- Batash R, Asna N, Schaffer P, Francis N, Schaffer M. Glioblastoma multiforme, diagnosis and treatment; recent literature review. Curr Med Chem. 2017;24:3002–9. - PubMed
-
- Sasmita AO, Wong YP, Ling APK. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol. 2018;14:40–51. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

