Divergent biological pathways distinguish community-acquired pneumonia from COVID-19 despite similar plasma cytokine profiles
- PMID: 40887575
- PMCID: PMC12400547
- DOI: 10.1186/s12931-025-03331-5
Divergent biological pathways distinguish community-acquired pneumonia from COVID-19 despite similar plasma cytokine profiles
Abstract
Background: Pulmonary infections, ranging from mild respiratory issues to severe multiorgan failure, pose a major global health threat. The immune response in community-acquired pneumonia (CAP) and COVID-19 influences disease severity and outcomes, but molecular pathogenesis differs across pathogens. Comparisons of plasma cytokine profiles between CAP and COVID-19 are limited. Analyzing these profiles with machine learning and bioinformatics could reveal subtle patterns and improve our understanding of immune responses in both conditions.
Methods: We conducted a novel case-control study to profile cytokine levels in patients with CAP and COVID-19. Age- and sex-matched cohorts included 39 patients with CAP, 39 with COVID-19, and 20 healthy controls. We measured 384 plasma cytokine levels using proximity extension assays and analyzed differences between cohorts with conventional statistical methods, bioinformatics and machine learning.
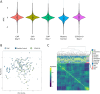
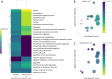
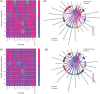
Results: Median ages of the cohorts were comparable (P = 0.797). COVID-19 patients exhibited a higher prevalence of hematologic disease (P = 0.047), increased corticosteroid use (P = 0.040), and reduced antibiotic use (P = 0.012). Clinical outcomes, including mortality, ICU admission, invasive mechanical ventilation, renal replacement therapy, acute respiratory distress syndrome, and acute kidney injury, were similar between groups. Both cohorts showed comparable absolute circulating cytokine profiles but distinct profiles relative to healthy controls. Machine learning identified a model of twelve cytokines that distinguished CAP from COVID-19 with a classification accuracy of 0.71 (SD 0.20). Gene ontology and enrichment analysis revealed differences in cytosolic and nuclear functions, intracellular signaling, stress responses, and cell cycle processes between patient cohorts and healthy controls. Enriched GO pathways showed that CAP pathways were positively associated with leukocyte counts and ARDS development, while COVID-19 pathways were negatively associated with ARDS and positively with platelet counts.
Conclusions: This case-control study provides insights into cytokine profiles related to CAP and COVID-19 pathogenesis. Although absolute circulating cytokine levels showed no significant differences between the groups, machine learning identified a model of twelve proteins that effectively distinguished the cohorts. Gene ontology and enrichment analyses also revealed distinct dysregulated pathways with differing associations with clinical variables in each cohort. These findings underscore the complexity and variability of cytokine responses in pulmonary infections.
Keywords: ARDS; Biomarkers; COVID-19; Community acquired pneumonia (CAP); Cytokines; Gene ontology; Machine learning; Proteomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Western University, Human Research Ethics Board (REB #s 6970; #100036; # 6963). Our experimental methods are performed in accordance with ethical standards of the responsible committee on human experimentation with the Helsinki Declaration of 1975. Informed written consent was obtained from either participants or their legal guardians. Consent for publication: Not applicable. Competing interests: Dr. Russell reports patents owned by the University of British Columbia (UBC) that are related to (1) the use of PCSK9 inhibitor(s) in sepsis, (2) the use of vasopressin in septic shock and (3) a patent owned by Ferring for use of selepressin in septic shock. Dr. Russell is an inventor on these patents. Dr. Russell is a shareholder in Molecular You Corp. Dr. Russell was a co-founder, Director and shareholder of Cyon Therapeutics Inc. (now closed) and Dr. Russell is currently a Cofounder, Director, and shareholder of Resolve Nanotherapeutics. Dr. Russell is the Senior Research Advisor of the British Columbia, Canada Post COVID – Interdisciplinary Clinical Care Network (PC-ICCN). Dr. Russell is no longer consulting for any industry. Dr. Russell reports receiving consulting fees in the last 3 years as a funded member of the Data and Safety Monitoring Board (DSMB) of an NIH-sponsored trial of plasma in COVID-19 (PASS-IT-ON) (2020–2021). Dr. Russell is the non-funded Chair of the Data and Safety Monitoring Board (DSMB) of a trial UC CISS II of stem cells in sepsis. Dr. Russell was a non-funded Science Advisor and member, Government of Canada COVID-19 Therapeutics Task Force (June 2020–2021). Dr. Russell has received grants for COVID-19 and for pneumonia research: 6 from the Canadian Institutes of Health Research (CIHR) and 3 from the St. Paul’s Foundation (SPF).
Figures



References
-
- Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, Cooley LA, Dean NC, Fine MJ, Flanders SA. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45–67. - PMC - PubMed
-
- Torres A, Cilloniz C, Niederman MS, Menendez R, Chalmers JD, Wunderink RG, van der Poll T. Pneumonia. Nat Rev Dis Primers. 2021;7(1):1–28. - PubMed
-
- Vos T, Lim SS, Abbafati C, Abbas KM, Abbasi M, Abbasifard M, Abbasi-Kangevari M, Abbastabar H, Abd-Allah F, Abdelalim A. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204–22. - PMC - PubMed
-
- Jeganathan N, Yau S, Ahuja N, Otu D, Stein B, Fogg L, Balk R. The characteristics and impact of source of infection on sepsis-related ICU outcomes. J Crit Care. 2017;41:170–6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

