DeepSCEM: A User-Friendly Solution for Deep Learning-Based Image Segmentation in Cellular Electron Microscopy
- PMID: 40888071
- PMCID: PMC12400132
- DOI: 10.1111/boc.70032
DeepSCEM: A User-Friendly Solution for Deep Learning-Based Image Segmentation in Cellular Electron Microscopy
Abstract
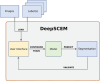
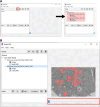
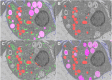
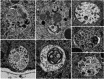
Deep learning methods using convolutional neural networks are very effective for automatic image segmentation tasks with no exception for cellular electron micrographs. However, the lack of dedicated easy-to-use tools largely reduces the widespread use of these techniques. Here we present DeepSCEM, a straightforward tool for fast and efficient segmentation of cellular electron microscopy images using deep learning with a special focus on efficient and user-friendly generation and training of models for organelle segmentation.
Keywords: cellular imaging; deep learning; electron microscopy; organelles; segmentation; software.
© 2025 The Author(s). Biology of the Cell published by Wiley‐VCH GmbH on behalf of Société Française des Microscopies and Société de Biologie Cellulaire de France.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Chaurasia, A. , and Culurciello E.. 2017. “LinkNet: Exploiting Encoder Representations for Efficient Semantic Segmentation.” 2017 IEEE Visual Communications and Image Processing (VCIP) , 1–4. 10.1109/VCIP.2017.8305148. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

