MCAMEF-BERT: an efficient deep learning method for RNA N7-methylguanosine site prediction via multi-branch feature integration
- PMID: 40889118
- PMCID: PMC12400811
- DOI: 10.1093/bib/bbaf447
MCAMEF-BERT: an efficient deep learning method for RNA N7-methylguanosine site prediction via multi-branch feature integration
Abstract
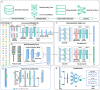
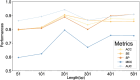
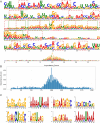
Accurate identification of N7-methylguanosine (m7G) modification sites plays a critical role in uncovering the regulatory mechanisms of various biological processes, including human development, tumor initiation, and progression. However, existing prediction methods still suffer from limited representational power, redundant feature fusion, insufficient utilization of biological prior knowledge, and poor interpretability. In this study, we propose a novel deep learning model named MCAMEF-BERT. This model adopts a parallel architecture that integrates both a DNABERT-2-based pretrained model branch and multiple traditional feature encoding branches, enabling comprehensive multi-perspective sequence feature extraction. To address the redundancy issue in feature fusion, we introduce a multi-channel attention module. Our model demonstrates superior accuracy and effectiveness on datasets from m7GHub, outperforming other state-of-the-art classifiers. Furthermore, we validate the interpretability of MCAMEF-BERT through in silico saturation mutagenesis experiments, and confirm its robustness in motif recognition. Moreover, its generalization capability is validated across diverse RNA modification site prediction tasks.
Keywords: in silico saturation mutagenesis experiments; DNABERT-2 pretrained model; N7-methylguanosine modification; multi-channel attention; multi-encoding fusion.
© The Author(s) 2025. Published by Oxford University Press.
Figures


Similar articles
-
Deep-m7G: A contrastive learning-based deep biological language model for identifying RNA N7-methylguanosine sites.Int J Biol Macromol. 2025 Jul;318(Pt 4):145341. doi: 10.1016/j.ijbiomac.2025.145341. Epub 2025 Jun 18. Int J Biol Macromol. 2025. PMID: 40553876
-
GenoM7GNet: An Efficient N7-Methylguanosine Site Prediction Approach Based on a Nucleotide Language Model.IEEE/ACM Trans Comput Biol Bioinform. 2024 Nov-Dec;21(6):2258-2268. doi: 10.1109/TCBB.2024.3459870. Epub 2024 Dec 10. IEEE/ACM Trans Comput Biol Bioinform. 2024. PMID: 39302806
-
iACP-DPNet: a dual-pooling causal dilated convolutional network for interpretable anticancer peptide identification.Funct Integr Genomics. 2025 Jul 4;25(1):147. doi: 10.1007/s10142-025-01641-x. Funct Integr Genomics. 2025. PMID: 40613943
-
Trajectory-Ordered Objectives for Self-Supervised Representation Learning of Temporal Healthcare Data Using Transformers: Model Development and Evaluation Study.JMIR Med Inform. 2025 Jun 4;13:e68138. doi: 10.2196/68138. JMIR Med Inform. 2025. PMID: 40465350 Free PMC article.
-
BBANsh: a deep learning architecture based on BERT and bilinear attention networks to identify potent shRNA.Brief Bioinform. 2025 Jul 2;26(4):bbaf443. doi: 10.1093/bib/bbaf443. Brief Bioinform. 2025. PMID: 40874819 Free PMC article.
References
-
- Huang H, Weng H, Deng X. et al. RNA modifications in cancer: functions, mechanisms, and therapeutic implications. Annual Review of Cancer Biology 2020;4:221–40. 10.1146/annurev-cancerbio-030419-033357 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

