Butyrate Reducing Bone Mass Loss by Regulating the Expression of m6A Methyltransferase METTL3 in Implant-Associated Staphylococcus aureus Osteomyelitis
- PMID: 40889211
- PMCID: PMC12401135
- DOI: 10.1111/jcmm.70683
Butyrate Reducing Bone Mass Loss by Regulating the Expression of m6A Methyltransferase METTL3 in Implant-Associated Staphylococcus aureus Osteomyelitis
Abstract
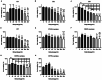
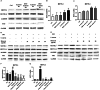
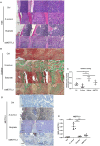
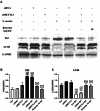
Staphylococcus aureus (S. aureus) has been identified as a hindrance to osteoblast differentiation, thereby contributing significantly to the development of osteomyelitis. Consequently, exploring pharmaceutical interventions targeting osteoblast differentiation mediated by S. aureus may present a novel approach for treating osteomyelitis. It has been reported that N6-methyladenosine (m6A) methylation is highly associated with infection. Moreover, studies continue to validate that short-chain fatty acids play an important role in transcriptional modification and have been considered as a potential treatment for S. aureus infection. Our research aimed to examine the impact and underlying mechanism of butyrate, a short-chain fatty acid, in reducing the inhibitory influence exerted by S. aureus on osteoblast differentiation. The concentration of butyrate beneficial to MC3T3-E1 cell viability was screened by Cell Counting Kit-8 (CCK8) assay. Reverse transcription-quantitative PCR (RT-qPCR), Western blotting, and alkaline phosphatase (ALP)staining were used to verify the osteogenic indexes and the expression levels of m6A methylation-related proteins in MC3T3-E1 cells infected with S. aureus at different time points. Besides, the same methods were used to verify the effects of butyrate stimulation and METTL3 knockdown on the osteogenic ability of MC3T3-E1 cells. H&E staining, Goldner staining, and immunohistochemical staining were used to verify the effect of butyrate on mice with endo-plant associated S. aureus osteomyelitis. The potential mechanisms of METTL3 and autophagy in MC3T3-E1 cells were studied by Western blotting. In vitro experiments, we found that butyrate significantly enhanced the expression of osteogenic-related genes down-regulated by infection in MC3T3-E1 cells induced by S. aureus, including RUNX2, OCN, and ALP. In addition, METTL3, an important m6A methyltransferase, was significantly up-regulated in S. aureus-infected MC3T3-E1 cells, but its expression could be down-regulated by butyrate. Inhibiting the expression of METTL3 by siRNA could effectively rescue the osteogenic markers down-regulated by S. aureus infection in MC3T3-E1 cells, which was similar to the results of butyrate treatment. In vivo experiments had shown that butyrate could alleviate inflammation and osteogenic activity in implant-associated osteomyelitis. The construction of bone marrow METTL3 low-expression mice using siRNA also showed similar effects on osteogenic activity. Additionally, Western blotting confirmed that knocking down METTL3 rescued the autophagy imbalance caused by S. aureus infection in MC3T3-E1 cells. In general, our research demonstrated that butyrate effectively alleviated the hindrance of osteoblast activity induced by S. aureus infection by suppressing the expression of METTL3, suggesting that butyrate may be a novel treatment for S. aureus osteomyelitis.
Keywords: Staphylococcus aureus; METTL3; N6‐methyladenosine; butyrate; osteomyelitis.
© 2025 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Knockout of the Staphylococcus aureus virulence gene sdrC promotes Myh7 expression to inhibit the progression of osteomyelitis.J Mol Histol. 2025 Jun 5;56(3):185. doi: 10.1007/s10735-025-10447-x. J Mol Histol. 2025. PMID: 40471432
-
[Mechanism of piR-hsa-26925 in regulating invasion and metastasis of lung adenocarcinoma via METTL3-mediated m6A methylation modification].Zhonghua Yi Xue Za Zhi. 2025 Sep 9;105(34):3002-3012. doi: 10.3760/cma.j.cn112137-20250121-00176. Zhonghua Yi Xue Za Zhi. 2025. PMID: 40904309 Chinese.
-
Resveratrol induces proliferation and differentiation of mouse pre-osteoblast MC3T3-E1 by promoting autophagy.BMC Complement Med Ther. 2023 Apr 14;23(1):121. doi: 10.1186/s12906-023-03943-8. BMC Complement Med Ther. 2023. PMID: 37060066 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

