Regulatory polymorphisms of MSH6, MSH2, FBXO11, and PPP1R21 genes affect survival of patients with immunotherapy-treated lung cancer
- PMID: 40889797
- PMCID: PMC12406856
- DOI: 10.1136/jitc-2025-011526
Regulatory polymorphisms of MSH6, MSH2, FBXO11, and PPP1R21 genes affect survival of patients with immunotherapy-treated lung cancer
Abstract
Background: Immune checkpoint inhibitors (ICI) improved survival of patients with non-small cell lung cancer (NSCLC), yet many patients do not respond to treatment. The identification of markers for ICI response remains an unmet clinical need. This study hypothesizes that host genetics influences the response to ICI, contributing to the variability in efficacy among individuals.
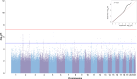
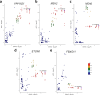
Methods: We conducted a genome-wide association study (GWAS) in patients with NSCLC on ICI monotherapy with nivolumab, pembrolizumab, or atezolizumab, to identify germline variants associated with objective response rate (ORR), progression-free survival (PFS), and overall survival (OS) at 24 months after the start of ICI therapy. Genomic DNA was genotyped using Axiom Precision Medicine Research Arrays. Raw data were processed with Axiom Analysis Suite, and quality checked with PLINK software. Imputation to the whole genome was done on the Michigan Imputation Server. Association analyses were performed for ORR (logistic regression with PLINK2 software) and survival (Cox proportional hazards model, with GenAbel package in R environment), with appropriate covariates. Variants were annotated for functional significance using SNPnexus and FUMA. Post-GWAS analyses, including colocalization, were performed to explore the function of the identified variants. Their possible role as expression quantitative trait loci was investigated in different databases (GTEx, eQTLGen, TCGA).
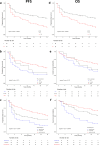
Results: No genome-wide significant associations were found for ORR or PFS, while a locus on chromosome 2 (lead variant: rs111648355) showed near genome-wide significance (p value=6.3×10⁻⁸) for OS. Patients with minor alleles of these variants exhibited significantly worse OS (HR=5.1, 95% CI: 2.9 to 9.2). Functional annotation linked these variants to regulatory effects on genes including MSH2, MSH6, PPP1R21, FBXO11, and STON1. These genes play a role in mismatch repair, endosomal trafficking, or major histocompatibility complex class II regulation, and might influence the response to immunotherapy.
Conclusions: This study identifies an association between a genomic locus on chromosome 2 and OS in patients with NSCLC treated with ICI. Although these results need validation in larger cohorts and functional studies to elucidate the underlying mechanisms, they highlight the potential of germline variants as predictive biomarkers of response to ICI.
Keywords: Genetic; Genome; Immune Checkpoint Inhibitors; Non-Small Cell Lung Cancer.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures
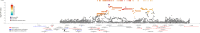
References
-
- Sezer A, Kilickap S, Gümüş M, et al. Cemiplimab monotherapy for first-line treatment of advanced non-small-cell lung cancer with PD-L1 of at least 50%: a multicentre, open-label, global, phase 3, randomised, controlled trial. The Lancet. 2021;397:592–604. doi: 10.1016/S0140-6736(21)00228-2. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
