Longitudinal liquid biopsy identifies an early predictive biomarker of immune checkpoint blockade response in head and neck squamous cell carcinoma
- PMID: 40890155
- PMCID: PMC12402333
- DOI: 10.1038/s41467-025-63538-4
Longitudinal liquid biopsy identifies an early predictive biomarker of immune checkpoint blockade response in head and neck squamous cell carcinoma
Abstract
Immune checkpoint blockade (ICB) has improved outcomes for patients with head and neck squamous cell carcinoma (HNSCC), but predictive biomarkers remain limited. Here, we use a time-resolved, multi-omic approach in a murine HNSCC model to characterize peripheral immune responses to ICB. Single-cell transcriptomics and T/B cell receptor analyses reveal early on-treatment expansion of effector memory T and B cell repertoires in responders, preceding tumor regression. These dynamic immune features inform a composite transcriptional signature that accurately predicts ICB response in independent human HNSCC cohorts. LiBIO outperforms existing biomarkers and generalizes to melanoma, non-small cell lung cancer, and breast cancer without retraining. These findings suggest that early treatment-induced changes in circulating immune repertoires reflect the host's capacity to mount an effective antitumor response. This work provides a framework for leveraging transient peripheral immune dynamics to develop non-invasive, high-fidelity biomarkers for response to immunotherapy across cancer types.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: E.R. is a co-founder of Medaware Ltd. ( https://www.medaware.com/ ), Metabomed ( https://www.metabomed.com/ ), and Pangea Biomed ( https://pangeamedicine.com/ ). He has divested and serves as an unpaid scientific consultant to the latter company. J.S.G. is a consultant/advisory board member for Pangea Biomed, Radionetics, and io9, and founder of Kadima Pharmaceuticals. The rest of the authors declare no conflicts of interest.
Figures
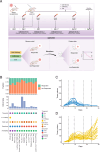



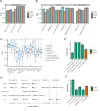
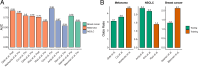
References
-
- Patel, S. P. & Kurzrock, R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther.14, 847–856 (2015). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

