AEBP1 drives fibroblast-mediated T cell dysfunction in tumors
- PMID: 40890191
- PMCID: PMC12402166
- DOI: 10.1038/s41467-025-63659-w
AEBP1 drives fibroblast-mediated T cell dysfunction in tumors
Abstract
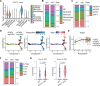
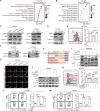
T cell dysfunction enables tumor immune evasion, understanding its mechanism is crucial for improving immunotherapy. Here we show, by RNA-sequencing analysis of human colon adenocarcinoma and triple-negative breast cancer tissues, that expression of Adipocyte Enhancer-Binding Protein 1 (AEBP1) positively correlates with T cell dysfunction and indicative of unfavorable patient outcomes. Subsequent single-cell RNA sequencing identifies cancer-associated fibroblasts (CAF) as the primary AEBP1 source. Fibroblast-specific AEBP1 deletion in mice enhances T cell cytotoxicity and suppresses tumor growth. Mechanistically, autocrine AEBP1 binds CKAP4 on CAFs, activating AKT/PD-L1 signaling to drive T cell dysfunction. By molecular-docking-based virtual screening we identify Chem-0199, a drug that disrupts the interaction between AEBP1 and CKAP4, thereby enhancing antitumor immunity. Both genetic and pharmacological AEBP1 inhibition synergize with immune checkpoint blockade in syngeneic models. Our study establishes AEBP1 as a key regulator of CAF-mediated T cell dysfunction and a therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Vinay, D. S. et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin. Cancer Biol.35, S185–S198 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

