Circadian disruption and ROS-NLRP3 signaling mediate sleep deprivation-enhanced silica nanoparticle toxicity in lacrimal glands
- PMID: 40890823
- PMCID: PMC12403269
- DOI: 10.1186/s12951-025-03630-5
Circadian disruption and ROS-NLRP3 signaling mediate sleep deprivation-enhanced silica nanoparticle toxicity in lacrimal glands
Abstract
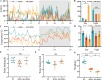
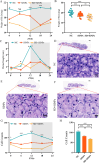
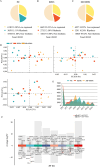
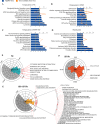
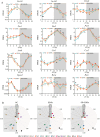
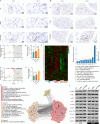
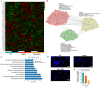
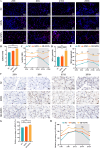
Sleep deprivation (SD) and exposure to engineered nanomaterials such as silica nanoparticles (SiNPs) are emerging risk factors for ocular surface disorders, particularly dry eye disease. However, the molecular mechanisms underlying their combined impact on lacrimal gland function remain unclear. In this study, we investigated the synergistic effects of SD and SiNPs exposure on circadian regulation, oxidative stress, inflammation, and structural integrity of the extraorbital lacrimal glands (ELGs) in C57BL/6J mice. Behavioral and physiological monitoring revealed that SD + SiNPs disrupted circadian locomotor activity and body temperature rhythms. Phenotypic assessments showed reduced tear secretion and ELG atrophy. RNA sequencing identified extensive transcriptomic reprogramming, including altered expression of core clock genes and enrichment of inflammatory and redox-related pathways. Increased reactive oxygen species (ROS) accumulation and γ-H2AX expression indicated oxidative DNA damage. Immunohistochemistry confirmed NLRP3 inflammasome activation, while Western blotting revealed enhanced phosphorylation of JAK2, STAT3, NF-κB p65, and IκBα, alongside upregulation of IL-17A. Elevated malondialdehyde levels further reflected oxidative lipid damage. These findings demonstrate that SD exacerbates SiNPs-induced ELG dysfunction via circadian disruption and activation of the ROS/NLRP3/IL-17A inflammatory axis. While these effects are currently limited to the lacrimal gland, future studies are needed to determine whether similar mechanisms contribute to broader systemic metabolic consequences.
Keywords: Circadian Rhythm Disruption; Environmental Nanotoxicology; Lacrimal Gland Dysfunction; NLRP3 Inflammasome; Oxidative Stress.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All experimental procedures were conducted following protocols approved by the Henan Provincial People’s Hospital’s Institutional Animal Care and Use Committee (HNEECA-2024-08), and were performed in accordance with ARRIVE guidelines. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Stark WJ, Stoessel PR, Wohlleben W, Hafner A. Industrial applications of nanoparticles. Chem Soc Rev. 2015;44:5793–805. - PubMed
-
- Dubey SK, Dey A, Singhvi G, Pandey MM, Singh V, Kesharwani P. Emerging trends of nanotechnology in advanced cosmetics. Colloid Surf B. 2022;214:112440. - PubMed
-
- Croissant JG, Butler KS, Zink JI, Brinker CJ. Synthetic amorphous silica nanoparticles: toxicity, biomedical and environmental implications. Nat Rev Mater. 2020;5:886–909.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

