PKP1 promotes lung cancer by modulating energy metabolism through stabilization of PFKP
- PMID: 40890861
- PMCID: PMC12403285
- DOI: 10.1186/s40364-025-00815-w
PKP1 promotes lung cancer by modulating energy metabolism through stabilization of PFKP
Abstract
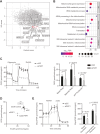
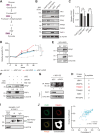
Lung cancer is the leading cause of cancer-related deaths worldwide, with lung squamous cell carcinoma (LUSC) lacking effective targeted therapies. Recent studies have identified Plakophilin-1 (PKP1) as one of the most differentially overexpressed genes in LUSC. This is particularly intriguing given that PKP1 is primarily known as a desmosomal component involved in cell adhesion, typically regarded as a tumor suppressor. To elucidate its biological role, we performed a genome-wide CRISPR knockout screening in PKP1-deficient models, revealing a strong dependence on mitochondrial metabolism. Metabolic assays further demonstrated that PKP1 loss significantly disrupts both mitochondrial function and glycolytic activity. In contrast, cells expressing PKP1 display a metabolically hyperactive phenotype, characterized by elevated oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). Building on these findings, we found that PKP1 depletion selectively reduces platelet-type phosphofructokinase (PFKP) levels, a key rate-limiting enzyme in glycolysis, by enhancing its ubiquitination and subsequent degradation. Functional rescue experiments confirmed that PFKP mediates the proliferative role of PKP1. These findings suggest that PKP1 overexpression in LUSC promotes a hyperactive metabolic state binding to TRIM21 and preventing PFKP degradation, facilitating tumor progression. These effects were consistently observed across multiple LUSC cell lines, underscoring the robustness of the mechanism. These findings highlight a potential therapeutic vulnerability in LUSC metabolic regulation.
Graphical Abstract:
Supplementary Information: The online version contains supplementary material available at 10.1186/s40364-025-00815-w.
Keywords: Glycolysis and OXPHOS; Phosphofructokinase; Plakophilin-1; TRIM21.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Reviews Clin Oncol 2023. 2023;20:9. - PubMed
-
- Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, et al. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355–64. - PubMed
-
- Angulo B, Suarez-Gauthier A, Lopez-Rios F, Medina PP, Conde E, Tang M, et al. Expression signatures in lung cancer reveal a profile for EGFR-mutant tumours and identify selective PIK3CA overexpression by gene amplification. J Pathol. 2008;214:347–56. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

