Defining the Antitumor Mechanism of Action of a Clinical-stage Compound as a Selective Degrader of the Nuclear Pore Complex
- PMID: 40891634
- PMCID: PMC12670082
- DOI: 10.1158/2159-8290.CD-25-0271
Defining the Antitumor Mechanism of Action of a Clinical-stage Compound as a Selective Degrader of the Nuclear Pore Complex
Abstract
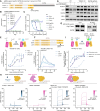
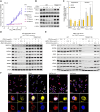
Cancer cells are acutely dependent on nuclear transport due to elevated transcriptional activity, suggesting an unrealized opportunity for selective therapeutic inhibition of the nuclear pore complex (NPC). Through large-scale phenotypic profiling of cancer cell lines, genome-scale functional genomic modifier screens, and mass spectrometry-based proteomics, we discovered that the clinical drug PRLX-93936 is a molecular glue that binds and reprograms the TRIM21 ubiquitin ligase to degrade the NPC. Upon compound-induced TRIM21 recruitment, the nuclear pore is ubiquitylated and degraded, resulting in the loss of short-lived cytoplasmic mRNA transcripts and the induction of cancer cell apoptosis. Direct compound binding to TRIM21 was confirmed via surface plasmon resonance and X-ray crystallography, whereas compound-induced TRIM21-nucleoporin complex formation was demonstrated through multiple orthogonal approaches in cells and in vitro. Phenotype-guided optimization yielded compounds with 10-fold greater potency and drug-like properties, along with robust pharmacokinetics and efficacy against pancreatic cancer xenografts and patient-derived organoids.
Significance: This study establishes the cancer therapeutic potential of optimized TRIM21 molecular glues to degrade the NPC and underscores the value of reexamining drugs with previously unknown mechanisms using current technologies.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
Stanford University has filed a patent application covering compounds in this manuscript (inventors: S.M. Corsello, L. Yuan, W. Ji, T. Zhang, N.S. Gray, and S.M. Hinshaw). L. Yuan reports grants from the Anderman Family Fellowship and the Stanford Medicine Dean’s Postdoctoral Fellowship during the conduct of the study. M.T. Tang reports grants from the NIH during the conduct of the study. C.J. Kuo reports grants from Chugai outside the submitted work and that he has a patent for tumor organoid culture issued and he is an Scientific Advisory Board member for Surrozen, Mozart, and NextVivo. N.S. Gray reports grants from the NIH, Stanford University, and the Michael J. Fox Foundation during the conduct of the study and personal fees from C4 Therapeutics, Lighthorse Therapeutics, Shenandoah, Matchpoint, Allorion Therapeutics, and Soltego outside the submitted work. S.M. Corsello reports grants from the Damon Runyon Cancer Research Foundation, the Shmunis Family Innovation Award in Cancer Therapeutics, the Stanford Innovative Medicines Accelerator, the NIH, and the c-ShaRP Voucher Program and other support from the Taylor Family Cancer Therapeutics Fund, the Novogradac Rivers Foundation, the Tortorici Family Pancreatic Cancer Research Fund, and the Lowell Berry Foundation during the conduct of the study as well as personal fees from Genentech outside the submitted work. No disclosures were reported by the other authors.
Figures





References
-
- Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817–25. - PubMed
-
- Di Marco T, Mazzoni M, Greco A, Cassinelli G. Non-oncogene dependencies: novel opportunities for cancer therapy. Biochem Pharmacol 2024;228:116254. - PubMed
-
- Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 2017;18:73–89. - PubMed
MeSH terms
Substances
Grants and funding
- S10OD030332/National Institutes of Health (NIH)
- P30 GM133894/GM/NIGMS NIH HHS/United States
- Lowell Berry Foundation
- S10 OD030332/OD/NIH HHS/United States
- T32GM139791/National Institutes of Health (NIH)
- R01 CA272485/CA/NCI NIH HHS/United States
- T32 GM139791/GM/NIGMS NIH HHS/United States
- R01CA272485/National Institutes of Health (NIH)
- School of Medicine Dean's Postdoctoral Fellowship
- Shmunis Family Innovation Award in Cancer Therapeutics
- CI 128-24/Damon Runyon Cancer Research Foundation (DRCRF)
- c-ShaRP Voucher Program
- K08 CA230220/CA/NCI NIH HHS/United States
- K08CA230220/National Institutes of Health (NIH)
- P30GM133894/National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Medical

