Discovery of potent and selective inhibitors of human NLRP3 with a novel mechanism of action
- PMID: 40892070
- PMCID: PMC12404154
- DOI: 10.1084/jem.20242403
Discovery of potent and selective inhibitors of human NLRP3 with a novel mechanism of action
Abstract
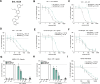
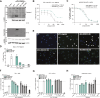
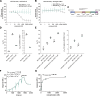
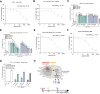
The NLRP3 inflammasome is an intracellular protein complex that causes inflammation via the release of IL-1β and pyroptosis. NLRP3 activation is associated with many age-related inflammatory diseases, and NLRP3 inhibition is a promising therapeutic strategy. We previously performed a DNA-encoded library screen to identify novel NLRP3-binding molecules. Herein we describe the characterization of BAL-0028 as a potent and specific inhibitor of NLRP3 signaling. Notably, BAL-0028 is a poor inhibitor of mouse NLRP3 but inhibits human and primate NLRP3 with nanomolar potency. Using cellular and biochemical analyses, we demonstrate that BAL-0028 binds to the NLRP3 NACHT domain at a site that is distinct from the MCC950-binding pocket. Using humanized NLRP3 mice, we show that a derivative of BAL-0028, BAL-0598, inhibits NLRP3 activation in vivo in a peritonitis model. Finally, we demonstrate that both BAL-0028 and BAL-0598 inhibit select hyperactive NLRP3 mutations associated with autoinflammatory diseases more potently than MCC950. BAL-0028 and BAL-0598 thus represent a new modality for NLRP3 inhibition in inflammatory diseases.
© 2025 Wilhelmsen et al.
Conflict of interest statement
Disclosures: K. Wilhelmsen, A. Deshpande, S. Tronnes, M. Banicki, M. Cochran, S. Cowdin, R. Montgomery, C.P. Portillo, P. Rubin, and Y. Wang reported grants and nonfinancial support from BioAge Labs, Inc. during the conduct of the study; and personal fees from BioAge Labs, Inc. outside the submitted work. M. Mahanta and T. Salazar reported personal fees and nonfinancial support from BioAge Labs, Inc. during the conduct of the study; and personal fees from BioAge Labs, Inc. outside the submitted work. K. Fortney and G. Hartman reported grants from BioAge Labs, Inc. during the conduct of the study; and personal fees from BioAge Labs, Inc. outside the submitted work. R.E. Hughes reported grants, personal fees, and nonfinancial support from BioAge Labs, Inc. outside the submitted work. M. Geyer reported grants and personal fees from BioAge Labs during the conduct of the study. R.C. Coll reported grants and personal fees from BioAge Labs during the conduct of the study; and personal fees from Viva in vitro diagnostics outside the submitted work. A.N.R. Weber is in the process of applying for a grant on NLRP3 research, but this has not influenced his contribution to this publication. In addition, K. Wilhelmsen, S. Tronnes, M. Mahanta, M. Banicki, M. Cochran, S. Cowdin, K. Fortney, R.E. Hughes, R. Montgomery, C.P. Portillo, P. Rubin, T. Salazar, and Y. Wang had patent numbers 11,702,391 and 11,708,334 issued (BioAge Labs, Inc.); B.F. Py reported a patent to PCT/EP2024/074525 issued; M. Geyer had a patent to 19/089,839 pending; and R.C. Coll had a patent to US 10,538,48 licensed, a patent to PCT/IB2017/053059 licensed, a patent to PCT/EP2017/053498 licensed, and a patent to US 18/906,535 pending. K. Wilhelmsen, A. Deshpande, S. Tronnes, M. Mahanta, M. Banicki, M. Cochran, S. Cowdin, K. Fortney, G. Hartman, R.E. Hughes, R. Montgomery, C.P. Portillo, P. Rubin, T. Salazar, and Y. Wang are employed by BioAge Labs, Inc., a biotechnology company developing NLRP3 inhibitors. M. Geyer and R.C. Coll are consultants for BioAge Labs, Inc. No other disclosures were reported.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

