Mechanism of Huanglian Wendan Decoction in ameliorating non-alcoholic fatty liver disease via modulating gut microbiota-mediated metabolic reprogramming and activating the LKB1/AMPK pathway
- PMID: 40892742
- PMCID: PMC12404375
- DOI: 10.1371/journal.pone.0331303
Mechanism of Huanglian Wendan Decoction in ameliorating non-alcoholic fatty liver disease via modulating gut microbiota-mediated metabolic reprogramming and activating the LKB1/AMPK pathway
Abstract
Background: Huanglian Wendan Decoction (HLWDD), a classical traditional Chinese medicine (TCM) formula, has shown therapeutic promise in treating metabolic disorders. However, its underlying mechanisms against non-alcoholic fatty liver disease (NAFLD) remain unclear.
Objective: This study aimed to elucidate the pharmacological mechanisms by which HLWDD ameliorates NAFLD, focusing on its impact on lipid metabolism, gut microbiota, and amino acid regulation.
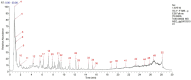
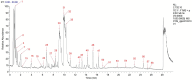
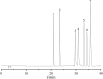
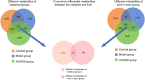
Methods: A NAFLD rat model was established by administering a high-sugar, high-fat, high-salt diet for 20 weeks. The core components of HLWDD were identified and quantified using UPLC-Q-TOF-MS/MS and HPLC, and further validated via network pharmacology and molecular docking. Therapeutic efficacy was assessed through analysis of body weight, serum lipid profiles, inflammatory cytokines, hepatic histology, and protein expression. Gut microbiota composition and liver-intestine metabolite profiles were evaluated using metagenomic sequencing and LC-MS/MS.
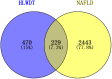
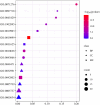
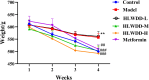
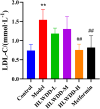
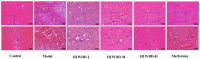
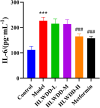
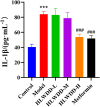
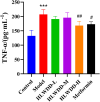
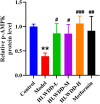
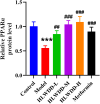
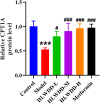
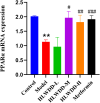
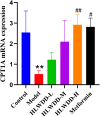
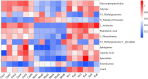
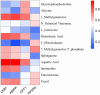
Results: Seven key constituents, including quercetin and berberine, were quantified (15.11-164.37 μg/mL) and shown to interact with lipid metabolism targets such as liver kinase B1 (LKB1), AMP-activated protein kinase (AMPK), peroxisome proliferator-activated receptor alpha (PPARα), and carnitine palmitoyltransferase 1A (CPT1A). HLWDD treatment significantly reduced body weight, hepatic lipid accumulation, and serum levels of triglycerides, total cholesterol, and low-density lipoprotein cholesterol, while increasing high-density lipoprotein cholesterol. Proinflammatory cytokines (IL-6, IL-1β, TNF-α) were notably suppressed. Mechanistically, HLWDD activated the LKB1/AMPK signaling pathway and modulated aspartic acid metabolism in association with increased abundance of Akkermansia in the gut. Metabolomic analysis identified 13 differential metabolites, with aspartic acid showing strong correlations with Akkermansia and LKB1/AMPK activity.
Conclusion: HLWDD exerts its anti-NAFLD effects by enhancing Akkermansia-mediated aspartate metabolism, thereby activating the LKB1/AMPK axis and promoting lipid oxidation via CPT1A and PPARα. This study provides new mechanistic insight into the gut-liver axis in NAFLD and highlights HLWDD as a multi-targeted therapeutic approach for restoring metabolic balance.
Copyright: © 2025 Zhu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
















Similar articles
-
Rhein alleviates hepatic steatosis in NAFLD mice by activating the AMPK/ACC/SREBP1 pathway to enhance lipid metabolism.Mol Med. 2025 Jul 10;31(1):255. doi: 10.1186/s10020-025-01304-4. Mol Med. 2025. PMID: 40640749 Free PMC article.
-
Hepatoprotective activity of mulberry extract in NAFLD mice for regulating lipid metabolism and inflammation identified via AMPK/PPAR-γ/NF-κB axis.J Ethnopharmacol. 2025 Jul 24;351:120108. doi: 10.1016/j.jep.2025.120108. Epub 2025 Jun 7. J Ethnopharmacol. 2025. PMID: 40490232
-
Uncovering the mechanisms of Zhubi decoction against rheumatoid arthritis through an integrated study of network pharmacology, metabolomics, and intestinal flora.J Ethnopharmacol. 2025 Jan 10;336:118736. doi: 10.1016/j.jep.2024.118736. Epub 2024 Aug 24. J Ethnopharmacol. 2025. PMID: 39186991
-
Gut Microbiota-Targeted Therapeutics for Metabolic Disorders: Mechanistic Insights into the Synergy of Probiotic-Fermented Herbal Bioactives.Int J Mol Sci. 2025 Jun 7;26(12):5486. doi: 10.3390/ijms26125486. Int J Mol Sci. 2025. PMID: 40564947 Free PMC article. Review.
-
Chinese Medicine for Non-Alcoholic Fatty Liver Disease: Targeting Key Signaling Pathways.Am J Chin Med. 2025;53(5):1379-1410. doi: 10.1142/S0192415X25500533. Epub 2025 Jul 18. Am J Chin Med. 2025. PMID: 40708491 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

