Transcriptomic changes associated with infection of Nicotiana benthamiana plants with tomato ringspot virus (genus Nepovirus) during the acute symptomatic stage and after symptom recovery
- PMID: 40892828
- PMCID: PMC12404439
- DOI: 10.1371/journal.pone.0328517
Transcriptomic changes associated with infection of Nicotiana benthamiana plants with tomato ringspot virus (genus Nepovirus) during the acute symptomatic stage and after symptom recovery
Abstract
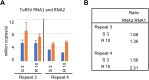
We have characterized the transcriptome of Nicotiana benthamiana plants infected with tomato ringspot virus (ToRSV), a nepovirus. We analyzed two different stages of infection: the acute systemic symptomatic stage and the symptom recovery stage in which young leaves emerge without visible symptoms. In agreement with previous observations, we note a similar concentration of viral RNAs in symptomatic and recovered leaves. Extensive reprogramming of the plant transcriptome was observed in symptomatic leaves, including upregulation of genes characteristic of biotic stress responses and downregulation of genes associated with the function and translation of chloroplasts. The majority of plant genes that were differentially regulated during the symptomatic stage returned to their basal levels after recovery. Thus, the extent of changes in the plant transcriptome was correlated with symptom intensity. However, we also identified genes that remained upregulated after the symptomatic stage or that were specifically induced at the symptom recovery stage. The list of genes that were upregulated at the symptom recovery stage was diverse and included several types of cysteine-rich antimicrobial peptides, notably two defensin-like genes that were specifically upregulated in recovered leaves, as confirmed by droplet-digital PCR. Several plant miRNAs were also differentially expressed in ToRSV-infected plants. Notably, miR391 was upregulated at both stages of infection, whereas miR530 and miR1919 were specifically upregulated during the symptomatic stage of infection. Several predicted miRNA targets were differentially regulated in our dataset, including new targets as well as previously validated targets (e.g., zinc finger A20/AN1 domain-containing stress-associated protein 1, a known target of miR530). Many of the miRNA predicted targets were related to plant defense responses and may contribute to symptom induction and/or symptom recovery.
Copyright: © 2025 Paudel et al. His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food, 2024. This material is licensed under the terms of the Canada Open Government Licence, which permits unrestricted use, distribution, and reproduction in any medium, provided you acknowledge the source of the Information or link to any attribution statement specified in the license. To view this licence, visit https://open.canada.ca/en/open-government-licence-canada.
Conflict of interest statement
The authors have declared that no competing interest exist. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Figures





References
-
- Sanfaçon H. Modulation of disease severity by plant positive-strand RNA viruses: The complex interplay of multifunctional viral proteins, subviral RNAs and virus-associated RNAs with plant signaling pathways and defense responses. Adv Virus Res. 2020;107:87–131. doi: 10.1016/bs.aivir.2020.04.003 - DOI - PubMed
-
- Osterbaan LJ, Fuchs M. Dynamic interactions between plant viruses and their hosts for symptom development. J Plant Pathol. 2019;101(4):885–95. doi: 10.1007/s42161-019-00323-5 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

