Optimizing blood-brain barrier permeability in KRAS inhibitors: A structure-constrained molecular generation approach
- PMID: 40893438
- PMCID: PMC12398849
- DOI: 10.1016/j.jpha.2025.101337
Optimizing blood-brain barrier permeability in KRAS inhibitors: A structure-constrained molecular generation approach
Abstract
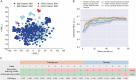
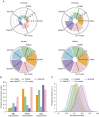
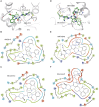
Kirsten rat sarcoma viral oncogene homolog (KRAS) protein inhibitors are a promising class of therapeutics, but research on molecules that effectively penetrate the blood-brain barrier (BBB) remains limited, which is crucial for treating central nervous system (CNS) malignancies. Although molecular generation models have recently advanced drug discovery, they often overlook the complexity of biological and chemical factors, leaving room for improvement. In this study, we present a structure-constrained molecular generation workflow designed to optimize lead compounds for both drug efficacy and drug absorption properties. Our approach utilizes a variational autoencoder (VAE) generative model integrated with reinforcement learning for multi-objective optimization. This method specifically aims to enhance BBB permeability (BBBp) while maintaining high-affinity substructures of KRAS inhibitors. To support this, we incorporate a specialized KRAS BBB predictor based on active learning and an affinity predictor employing comparative learning models. Additionally, we introduce two novel metrics, the knowledge-integrated reproduction score (KIRS) and the composite diversity score (CDS), to assess structural performance and biological relevance. Retrospective validation with KRAS inhibitors, AMG510 and MRTX849, demonstrates the framework's effectiveness in optimizing BBBp and highlights its potential for real-world drug development applications. This study provides a robust framework for accelerating the structural enhancement of lead compounds, advancing the drug development process across diverse targets.
Keywords: Blood-brain barrier permeability; Deep learning; Drug design; Generation models; KRAS inhibitors; Molecular optimization.
© 2025 The Authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
-
- Patricelli M.P., Janes M.R., Li L.-S., et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. - PubMed
-
- Lanman B.A., Allen J.R., Allen J.G., et al. Discovery of a covalent inhibitor of KRASG12C (AMG 510) for the treatment of solid tumors. J. Med. Chem. 2020;63:52–65. - PubMed
-
- Fell J.B., Fischer J.P., Baer B.R., et al. Identification of the clinical development candidate MRTX849, a covalent KRASG12C inhibitor for the treatment of cancer. J. Med. Chem. 2020;63:6679–6693. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

