Exploring omics strategies for drug discovery from Actinomycetota isolated from the marine ecosystem
- PMID: 40894205
- PMCID: PMC12394500
- DOI: 10.3389/fphar.2025.1634207
Exploring omics strategies for drug discovery from Actinomycetota isolated from the marine ecosystem
Abstract
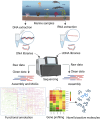
Marine Actinomycetota are prolific producers of diverse bioactive secondary metabolites, making them vital for drug discovery. Traditional cultivation and bioassay-guided isolation techniques often lead to the rediscovery of the same compounds, revealing the limitations of these traditional approaches and emphasizing the need for more advanced methods. The emergence of omics technologies such as genomics, metagenomics, transcriptomics, and metabolomics has dramatically enhanced the ability to investigate microorganisms by providing detailed insights into their biosynthetic gene clusters, metabolic pathways, and regulatory mechanisms. These comprehensive tools facilitate the discovery and functional analysis of new bioactive compounds by revealing the genetic blueprints underlying their biosynthesis. Omics and function-driven techniques like heterologous expression, analytical techniques (including high-resolution mass spectrometry and nuclear magnetic resonance spectroscopy), and culture condition optimization have enabled access to previously silent or cryptic gene clusters, expanding the chemical diversity available for exploration. This review emphasizes the integration of omics-based insights with function-driven methodologies and innovative culture techniques, forming a holistic approach to unlock the extensive biosynthetic capabilities of marine Actinomycetota. Combining these strategies holds great promise for discovering new marine-derived compounds with potential therapeutic applications.
Keywords: culture-independent analysis; drug discovery; marine Actinomycetota; marine ecosystem; omics approaches.
Copyright © 2025 Narsing Rao, Quadri, Sathish, Quach, Li and Thamchaipenet.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
References
-
- Adnani N., Chevrette M. G., Adibhatla S. N., Zhang F., Yu Q., Braun D. R., et al. (2017). Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, Keyicin. ACS Chem. Biol. 12 (12), 3093–3102. 10.1021/acschembio.7b00688 - DOI - PMC - PubMed
-
- Almeida E. L., Carrillo Rincón A. F., Jackson S. A., Dobson A. D. W. (2019). Comparative Genomics of marine sponge-derived Streptomyces spp. Isolates SM17 and SM18 with their closest terrestrial relatives provides novel insights into environmental niche adaptations and secondary metabolite biosynthesis potential. Front. Microbiol. 10, 1713. 10.3389/fmicb.2019.01713 - DOI - PMC - PubMed
-
- Amos G. C. A., Awakawa T., Tuttle R. N., Letzel A. C., Kim M. C., Kudo Y., et al. (2017). Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc. Natl. Acad. Sci. U. S. A. 114 (52), E11121-E11130–E11130. 10.1073/pnas.1714381115 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources