This is a preprint.
Single-Assay Characterization of Ternary Complex Assembly and Activity in Targeted Protein Degradation
- PMID: 40894543
- PMCID: PMC12393538
- DOI: 10.1101/2025.08.20.671298
Single-Assay Characterization of Ternary Complex Assembly and Activity in Targeted Protein Degradation
Abstract
Targeted protein degradation (TPD) is a rapidly advancing therapeutic strategy that selectively eliminates disease-associated proteins by co-opting the cell's protein degradation machinery. Covalent modification of proteins with ubiquitin is a critical event in TPD, yet the analytical tools for quantifying the ubiquitination kinetics have been limited. Here, we present a real-time, high-throughput fluorescent assay utilizing purified, FRET-active E2-Ub conjugates to monitor ubiquitin transfer. This assay is highly versatile, requiring no engineering of the target protein or ligase, thereby accelerating assay development and minimizing the risk of artifacts. The single-step, single-turnover nature of the monitored reaction enables rigorous and quantitative analysis of ubiquitination kinetics. We show that this assay can be used to measure key degrader characteristics such as degrader affinity for the target protein, degrader affinity for the ligase, affinity of ternary complex assembly, and catalytic efficiency of the ternary complex. The high sensitivity and accuracy of this comprehensive, single-assay approach to ternary complex characterization will empower the discovery and optimization of heterobifunctional degraders and molecular glues.
Conflict of interest statement
Competing interests Pending patent applications PCT/US2022/048268, US 18/712304 and EU 22899266.5 list the Board of Regents, The University of Texas System as the applicant and D.N.I. as an inventor. D.N.I. is a co-founder and a shareholder of E3 Bioscience LLC, a commercial entity that manufactures reagents described in this study. All other authors declare no competing interests.
Figures




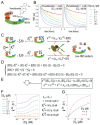
References
-
- Komander D. & Rape M. The ubiquitin code. Annu Rev Biochem 81, 203–29 (2012). - PubMed
-
- Berndsen C.E. & Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 21, 301–7 (2014). - PubMed
-
- Scott D.C. & Schulman B.A. Dual-color pulse-chase ubiquitination assays to simultaneously monitor substrate priming and extension. Methods Enzymol 618, 29–48 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
