This is a preprint.
Conformational dynamics underlying slow inactivation in voltage-gated sodium channels
- PMID: 40894622
- PMCID: PMC12393289
- DOI: 10.1101/2025.08.14.670348
Conformational dynamics underlying slow inactivation in voltage-gated sodium channels
Abstract
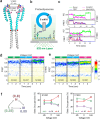
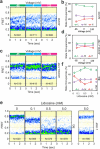
Voltage-gated sodium (Nav) channels initiate and propagate action potentials in many excitable cells. Upon repetitive activation, the fraction of Nav channels available for excitation gradually decreases on a timescale ranging from seconds to minutes, a phenomenon known as slow inactivation. This process is crucial for regulating cellular excitability and firing patterns. Slow inactivation is proposed to result from the collapse of the selectivity filter pore coupled with the opening of the primary helix bundle crossing gate. However, the conformational changes underlying slow inactivation and the molecular coupling between the selectivity filter and primary gate remain unclear. In this study, we investigated the conformational dynamics of the selectivity filter in prokaryotic NavAb channels reconstituted into liposomes using single-molecule FRET (smFRET). Our smFRET data revealed the conformational transitions in the NavAb selectivity filter pore among three distinct states, with activating voltages enriching the high-FRET conformations, potentially associated with slow inactivation. Using electrophysiological and crystallographic methods, we further identified the L176 residue in the selectivity filter P1 helix as a critical coupler between the primary and slow inactivation gates. We showed that L176 mutations with side chains of larger sizes significantly facilitated the slow inactivation of the NavAb channel, and the L176F mutation forced the opening mutant carrying the C-terminal deletion to be crystallized at the closed state. Consistently, our smFRET results revealed that C-terminal deletion markedly attenuated the high FRET conformation of the selectivity filter, which was restored by the L176F mutation. Moreover, using the classical Nav open-pore blocker lidocaine, we showed that it also depleted the high FRET conformation of the NavAb selectivity filter in a dose-dependent manner. The L176F mutation, again, markedly reversed the conformational shifts caused by lidocaine, an effect similar to it on the opening mutant carrying the C-terminal deletion. Our studies consistently suggested that slow inactivation in the NavAb channel is underlined by the collapse of the selectivity filter pore, represented by the high FRET conformation uncovered by our smFRET measurements, while the L176 residue at the P1 helix of the selectivity filter and T206 at the pore lining helix couple the conformational changes of the slow inactivation gate at selectivity filter and the primary gate at the helix bundle crossing.
Figures






Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Structural basis of fast N-type inactivation in Kv channels.Nature. 2025 Aug 6. doi: 10.1038/s41586-025-09339-7. Online ahead of print. Nature. 2025. PMID: 40770100
-
Electrophoresis.2025 Jul 14. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 14. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36251838 Free Books & Documents.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
References
-
- Hille B. Ion channels of excitable membranes, xviii, 814 p. (Sinauer Associates Inc., Sunderland, MA USA, 2001).
-
- de Lera Ruiz M. & Kraus R.L. Voltage-Gated Sodium Channels: Structure, Function, Pharmacology, and Clinical Indications. J Med Chem 58, 7093–118 (2015). - PubMed
-
- Huang J., Pan X. & Yan N. Structural biology and molecular pharmacology of voltage-gated ion channels. Nat Rev Mol Cell Biol 25, 904–925 (2024). - PubMed
-
- Catterall W.A., Lenaeus M.J. & Gamal El-Din T.M. Structure and Pharmacology of Voltage-Gated Sodium and Calcium Channels. Annu Rev Pharmacol Toxicol 60, 133–154 (2020). - PubMed
-
- Ulbricht W. Sodium channel inactivation: molecular determinants and modulation. Physiol Rev 85, 1271–301 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
