This is a preprint.
Macrophages drive a fibrogenic gene program of periductal fibroblasts in pediatric primary sclerosing cholangitis
- PMID: 40894696
- PMCID: PMC12393299
- DOI: 10.1101/2025.08.14.670195
Macrophages drive a fibrogenic gene program of periductal fibroblasts in pediatric primary sclerosing cholangitis
Abstract
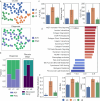
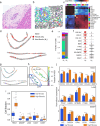
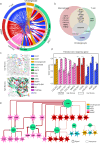
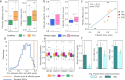
Primary sclerosing cholangitis (PSC) is an autoimmune, cholestatic liver disease characterized by inflammation and fibrosis surrounding bile ducts. The cellular crosstalk driving periductal fibrosis remains poorly defined. This study applied a multi-omics approach integrating MERSCOPE spatial transcriptomics, bulk RNA-seq, and SomaScan proteomics to characterize fibrotic periductal regions and their cell-cell communications. Macrophages (MP) expressing moderate-to-high CD163 were found co-localized with cholangiocytes, T cells, and collagen-producing hepatic stellate cells (HSC). Cell niche analysis identified periductal regions with elevated fibrotic signals, in which cell-cell communication analysis revealed MP-HSC interactions involving 17 fibrotic driver genes in MP (e.g., IFNGR1, CSF1R, CD163) and six fibrotic effector genes in HSC (e.g., COL1A2, VCAN, MMP2). In validation analyses, bulk RNA-seq data showed higher driver and effector gene scores in PSC with established fibrosis compared to early-stage PSC and autoimmune hepatitis (AIH). Plasma proteins encoded by MP driver genes were elevated in autoimmune liver disease (AILD) and in patients with elevated (≥3.29 kPa) liver stiffness on MR elastography. These findings suggest that macrophages engage in localized crosstalk with HSC, activating fibrotic gene programs and contributing to periductal fibrosis in PSC, thereby identifying potential molecular targets for therapeutic intervention.
Keywords: Primary sclerosing cholangitis; cell-cell communication; macrophage; periductal fibrosis; spatially resolved transcriptomics.
Figures


References
-
- Deneau MR, El-Matary W, Valentino PL, Abdou R, Alqoaer K, Amin M, et al. The natural history of primary sclerosing cholangitis in 781 children: A multicenter, international collaboration. Hepatology. 2017;66(2):518–27. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
