This is a preprint.
Visualizing Poloidal Orientation in DNA Minicircles
- PMID: 40894713
- PMCID: PMC12393517
- DOI: 10.1101/2025.08.20.671369
Visualizing Poloidal Orientation in DNA Minicircles
Abstract
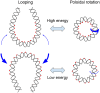
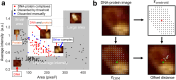
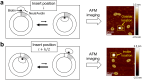
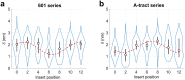
A short (<150 bp) double-stranded DNA (dsDNA) molecule ligated end-to-end forms a DNA minicircle. Due to sequence-dependent, nonuniform bending energetics, such a minicircle is predicted to adopt a certain inside-out orientation, known as the poloidal orientation. Despite theoretical and computational predictions, experimental evidence for this phenomenon has been lacking. In this study, we introduce a single-molecule approach to visualize the poloidal orientation of DNA minicircles. We constructed a set of DNA minicircles, each containing a single biotin located at a different position along one helical turn of the dsDNA, and imaged the location of biotin-bound NeutrAvidin relative to the DNA minicircle using atomic force microscopy (AFM). We applied this approach to two DNA sequences previously predicted to exhibit strongly preferred poloidal orientations. The observed relative positions of NeutrAvidin shifted between the inside and outside of the minicircle with different phases, indicating distinct poloidal orientations for the two sequences. Coarse-grained simulations revealed narrowly distributed poloidal orientations with different mean orientations for each sequence, consistent with the AFM results. Together, our findings provide experimental confirmation of preferred poloidal orientations in DNA minicircles, offering insights into the intrinsic dynamics of circular DNA.
Conflict of interest statement
DECLARATION OF INTEREST The authors declare no competing interests.
Figures


References
-
- Ma N. and van der Vaart A., Anisotropy of b-dna groove bending, Journal of the American Chemical Society 138, 9951 (2016). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
