Identification of superior rice donors with enhanced nitrogen use efficiency using a comprehensive multivariate genotype selection strategy
- PMID: 40894902
- PMCID: PMC12398238
- DOI: 10.1016/j.isci.2025.113280
Identification of superior rice donors with enhanced nitrogen use efficiency using a comprehensive multivariate genotype selection strategy
Abstract
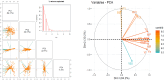
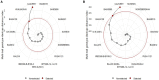
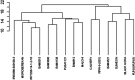
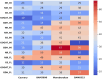
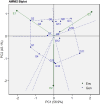
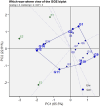
Improving nitrogen use efficiency (NUE) of rice plants utilizing a few end-of-season traits poses a severe phenotyping bottleneck in exploring the genetic diversity of a large population and genotype selection accuracy. Therefore, a comprehensive multivariate genotype selection strategy was developed to explore maximum genetic variation of 300 diverse rice genotypes and accurately select promising rice donors with enhanced NUE traits on a multi-year (2019, 2021, and 2022) -trait (126 traits) -environment (2) -temporal (5) -location (3) scale. The multi-trait genotype ideotype distance index (MGIDI) ranked Cauvery, Suweon, RPW9-4 (SSI) and BAM3690 (IC463705) as superior NUE genotypes; Moroberekan, PUSA1121 and BAM8315 (Basmati 370) as low NUE genotypes. The multi-location field performance and molecular analysis of key nitrogen assimilatory genes confirmed the outperformance of the Cauvery genotype in terms of possessing efficient N sensing, uptake, transport and assimilation characteristics under N-limited conditions. Phenome-wide multivariate analysis highlights root-shoot plastic response as a key target trait for breeding rice genotypes resilient to N stress conditions.
Keywords: Agricultural science; Genomic library; Genotyping; Natural sciences; Plant biology.
© 2025 Published by Elsevier Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- FAOSAT . FAO Statistical Database; 2022. Food and Agricultural Organization of the United Nation.https://www.fao.org/faostat/en/#data/QCL
-
- Elangovan A., Duc N.T., Raju D., Kumar S., Singh B., Vishwakarma C., Gopala Krishnan S., Ellur R.K., Dalal M., Swain P., et al. Imaging sensor-based high-throughput measurement of biomass using machine learning models in rice. Agriculture. 2023;13:852.
LinkOut - more resources
Full Text Sources

