Genetic determinants of testicular sperm extraction outcomes: insights from a large multicentre study of men with non-obstructive azoospermia
- PMID: 40896145
- PMCID: PMC12396851
- DOI: 10.1093/hropen/hoaf049
Genetic determinants of testicular sperm extraction outcomes: insights from a large multicentre study of men with non-obstructive azoospermia
Abstract
Study question: What is the diagnostic yield and the pre-testicular sperm extraction (TESE) prognostic value of a non-obstructive azoospermia (NOA)-specific virtual gene panel?
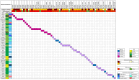
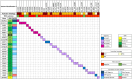
Summary answer: The diagnostic yield in our cohort was 6.1%, and by combining our data with published literature, we identified 11 genes compatible with testicular sperm production and 19 genes associated with no sperm retrieval in carriers of pathogenic (P) or likely pathogenic (LP) mutations.
What is known already: Azoospermia, the most severe form of male infertility, affects ∼1% of the male population, with TESE being the primary treatment option. However, in NOA, TESE fails in nearly 50% of cases and existing clinical parameters are unable to predict TESE failure. Over the past decade, next-generation sequencing (NGS) has identified several candidate NOA genes, but their diagnostic utility and impact on TESE outcomes have not been fully explored.
Study design size and duration: A literature search was addressed to identify well-established NOA genes for designing a specific virtual gene panel for NOA. Our retrospective study analysed the diagnostic yield of the NGS-based virtual gene panel, comprising 145 genes, in 571 men affected by idiopathic NOA with known TESE outcomes. Subsequently, a second literature search was performed to identify carriers of LP/P variants in the genes where we identified mutations, focusing on individuals with known TESE outcomes. This approach allowed us to integrate the published data with our findings and predict a genotype-phenotype correlation between the affected genes and TESE success.
Participants/materials settings methods: 571 NOA patients with known TESE outcomes were recruited in two European and one Middle East centres. Variants were obtained from a whole-exome sequencing dataset and crossed with the 145 genes of the virtual gene panel. After a filtering process, variants were manually assessed and classified according to ACMG guidelines by using two methods: (i) In order to compare our data with previously published studies, we applied ACMG-AMP guidelines along with ClinGen recommendations used by other similar studies. (ii) A new approach was used to optimize ACMG-AMP guidelines with all ClinGen recommendations and incorporated NOA-specific rules addressing phenotypic, locus, and allelic heterogeneity. LP and P variants were confirmed by Sanger sequencing.
Main results and the role of chance: By using the new variant classification approach adapted for NOA, we identified LP/P variants in 6.1% of patients, with a higher yield (9.4%) in cases with negative TESE outcomes and maturation arrest (11.7%). By integrating our findings with the literature, we highlight 19 genes recurrently associated with negative TESE outcomes and 11 genes associated with positive sperm retrieval either in the testis or in semen. TESE is recommended for patients with LP or P variants in the 11 specific genes. Notably, six of these genes are located on the X chromosome, therefore, these variants will be obligatorily transmitted to daughters, and potentially increase the risk of NOA-related infertility in male offspring. We observed that nine genes, in which we identified LP/P variants, have been previously described in individuals with premature ovarian insufficiency (POI). Of these, eight were associated with negative TESE outcomes in men. Furthermore, we propose seven additional genes mutated in our cohort of NOA patients as novel POI candidates. These genes have not yet been considered as POI candidates, but they result in female infertility when knocked out in mouse models.
Large scale data: LP/P variants have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Limitations reasons for caution: NOA is genetically heterogeneous, and our panel excludes those genes which were reported only in a single subject or single family. Although this can limit the diagnostic yield in our study, it ensures that only genes with clear relationship with NOA have been analysed. While in our cohort TESE outcomes are known for all patients, this information is often not available for mutation carriers in the published studies. Consequently, the total number of patients with P variants in the same gene remains relatively low, limiting our final conclusions. However, even if the number of carriers of genes associated with positive sperm retrieval is relatively low, it does not constrain our conclusions regarding TESE prediction. On the other hand, caution is warranted for genes linked to negative TESE outcomes, except for TEX11, SYCE1, and MSH4, each of which have 10 or more reported TESE-negative cases.
Wider implications of the findings: Our study was performed on the largest available NOA cohort with known TESE outcomes. It not only provides an estimate on the diagnostic potential of a NOA-specific virtual gene panel, but it also advances the understanding of genetic factors influencing TESE outcomes. Half of the genes mutated in our study and presenting TESE-positive outcomes are already informative for clinical decision-making. The observed genotype-phenotype correlations may help in personalized decision-making prior to TESE, in order to undergo the procedure or to avoid unnecessary invasive treatment. It provides valuable insights that can inform clinical management strategies and potentially offer personalized treatments based on genetic profiles. The use of two different variant classification methods highlights that previous studies may have over-estimated the diagnostic yield, underscoring the need for a standardized variant classification approach addressed specifically to male infertility. Our study also emphasizes the overlap between NOA- and POI-associated genes, which has important clinical implications for genetic counselling of female siblings of affected individuals.
Study funding/competing interests: This work was funded by the Spanish Ministry of Health Instituto Carlos III-FIS FONDOS FEDER (grant numbers PI20/01562 and PI23/00425) and the Fanconi Research Fund awarded to C.K. and A.R.-E. This article is based upon work from COST Action CA20119 (ANDRONET), supported by COST (European Cooperation in Science and Technology) (www.cost.eu). C.K., A.R.-E., G.F., M.J.X., M.S.O., C.A., M.S., and E.R.-C. are members of the Action. This research was also supported by the Qatar National Research Fund (QNRF) under grant NPRP12S-0318-190394, and by an Investigator Award in Science from the Wellcome Trust (209451 to J.A.V.). The authors declare no competing interests.
Keywords: POI; TESE; azoospermia; diagnostic; exome; gene panel; genetics; male infertility; spermatogenesis.
© The Author(s) 2025. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology.
Conflict of interest statement
None declared.
Figures


References
-
- Araujo TF, Friedrich C, Grangeiro CHP, Martelli LR, Grzesiuk JD, Emich J, Wyrwoll MJ, Kliesch S, Simões AL, Tüttelmann F. Sequence analysis of 37 candidate genes for male infertility: challenges in variant assessment and validating genes. Andrology 2020;8:434–441. - PubMed
-
- Batiha O, Burghel GJ, Alkofahi A, Alsharu E, Smith H, Alobaidi B, Al-Smadi M, Awamlah N, Hussein L, Abdelnour A et al. Screening by single-molecule molecular inversion probes targeted sequencing panel of candidate genes of infertility in azoospermic infertile Jordanian males. Hum Fertil (Camb) 2022;25:939–946. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
