Haplotype shifts in the lipid-related OsGELP gene family underpin rice adaptation to high latitudes
- PMID: 40897734
- PMCID: PMC12405586
- DOI: 10.1038/s41598-025-15488-6
Haplotype shifts in the lipid-related OsGELP gene family underpin rice adaptation to high latitudes
Abstract
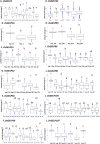
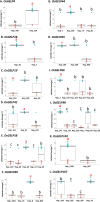
Rice (Oryza sativa L.) originated in tropical regions but has successfully adapted to higher latitudes, largely through modifications in photoperiod sensitivity and cold tolerance mechanisms. Lipid metabolism, particularly involving membrane-associated enzymes, plays a key role in environmental sensing and response. The GDSL esterase/lipase (GELP) gene family, known for its lipid-related enzymatic activity, has been associated with various abiotic stress responses. In this study, we investigated the potential role of OsGELP genes in rice adaptation to high-latitude environments using publicly available genomic and geolocation data from 3,000 rice accessions. We identified haplotypes for all 115 OsGELP genes and classified them based on single-nucleotide polymorphisms (SNPs) and the mean latitude of occurrence. Haplotypes with average latitudinal distribution above 35°N were defined as high-latitude haplotypes (HLHs). We selected 10 OsGELP genes containing 11 HLHs for further analysis. Haplotype network and amino acid sequence analyses revealed that a limited number of non-synonymous mutations in HLHs may have conferred adaptive advantages to high-latitude environments. Notably, these HLH-associated OsGELP genes are predominantly expressed in roots, suggesting a root-mediated mechanism of environmental adaptation. Interestingly, only 3 of 14 known cold tolerance genes in rice exhibited HLHs, highlighting the distinct role of OsGELP genes in latitude adaptation. Our findings suggest that these 10 OsGELP genes with HLHs may represent previously unrecognized genetic components contributing to high-latitude adaptation in rice.
Keywords: OsGELP; Adaptation; GDSL-type esterase/lipase; High-latitude haplotype; Rice; Root.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

