Chloroplast genome comparison and taxonomic reassessment of Polygonatum sensu Lato (Asparagaceae): implications for molecular marker development in traditional medicinal plants
- PMID: 40898047
- PMCID: PMC12403319
- DOI: 10.1186/s12864-025-12012-y
Chloroplast genome comparison and taxonomic reassessment of Polygonatum sensu Lato (Asparagaceae): implications for molecular marker development in traditional medicinal plants
Abstract
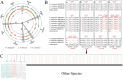
The Polygonati Rhizoma have generated significant market attention for their medicinal and culinary applications. However, morphological similarities and ambiguous species boundaries complicate the identification of genera and species, thereby impeding product development and utilization within Polygonatum sensu lato. Despite the widespread application of the chloroplast genome for taxonomic boundary revisions for Polygonatum s.l., a critical gap persist regarding their genomic applicability and the lack of standardized pipelines for developing species-specific molecular markers capable of rapid discrimination among species. This study aims to assess the effectiveness of chloroplast genomes in clarifying the current taxonomic status of the genera and species of Polygonatum s.l., and develop a reliable process for rapid identification of designated species from other species. A total of 21 chloroplast genomes were sequenced and assembled, and subsequent analyses included phylogenetic inference, multiple molecular species delimitation methods, and an automated screening framework were employed for subsequent analysis. Comparative analyses revealed relatively conserved chloroplast genomes, with notable variation limited primarily to the length of IR and LSC regions. By integrating multiple delimitation methods, the chloroplast genome validated 82.46% of the current classifications of Polygonatum s.l., demonstrating strong support (90.63%) for species represented by multiple sequences, yet only moderate support (70%) for those with single-sequence representation. Additionally, this study established and validated a scalable molecular marker development framework, spanning from identification of species-specific SNPs/InDels to the design of high-resolution molecular markers, illustrated through case studies involving Heteropolygonatum and three medicinally significant Polygonatum species.
Keywords: Heteropolygonatum; DNA barcoding; Molecular marker; Polygonati rhizoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Complete mitochondrial genome of Polygonatum cyrtonema Hua reveals variation diversity and evolutionary trends.BMC Plant Biol. 2025 Aug 21;25(1):1109. doi: 10.1186/s12870-025-07074-9. BMC Plant Biol. 2025. PMID: 40841885 Free PMC article.
-
Comparative analysis of 18 chloroplast genomes reveals genomic diversity and evolutionary dynamics in subtribe Malaxidinae (Orchidaceae).BMC Plant Biol. 2025 Aug 2;25(1):1013. doi: 10.1186/s12870-025-06772-8. BMC Plant Biol. 2025. PMID: 40753185 Free PMC article.
-
Comparative Analysis of Chloroplast Genomes of 19 Saxifraga Species, Mostly from the European Alps.Int J Mol Sci. 2025 Jun 23;26(13):6015. doi: 10.3390/ijms26136015. Int J Mol Sci. 2025. PMID: 40649794 Free PMC article.
-
Harnessing Molecular Phylogeny and Chemometrics for Taxonomic Validation of Korean Aromatic Plants: Integrating Genomics with Practical Applications.Plants (Basel). 2025 Aug 1;14(15):2364. doi: 10.3390/plants14152364. Plants (Basel). 2025. PMID: 40805713 Free PMC article. Review.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
References
-
- Chen XQ, Tamura MN. Polygonatum. In: Wu ZY, Peter PH, editors. Flora of China. Beijing: Science; St. Louis: Missouri Botanical Garden Press;; 2000. pp. 223–32.
-
- Floden AJ, Schilling EE. Using phylogenomics to reconstruct phylogenetic relationships within tribe polygonateae (Asparagaceae), with a special focus on Polygonatum. Mol Phylogenet Evol. 2018;129:202–13. - PubMed
-
- Jeffrey C. The genus Polygonatum (Liliaceae) in Eastern Asia. Kew Bull. 1980;34:435–71.
-
- Jeffrey C. Further note on Eastern Asian Polygonatum (Liliaceae). Kew Bull. 1982;37:335–9.
-
- Tamura MN, Ogisu M, Xu J. Heteropolygonatum, a new genus of the tribe polygonateae (Convallariaceae) from West China. Kew Bull. 1997;52:949–56.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

