Improving immune-related health outcomes post-cesarean birth with a gut microbiome-based program: A randomized controlled trial
- PMID: 40898384
- PMCID: PMC12405607
- DOI: 10.1111/pai.70182
Improving immune-related health outcomes post-cesarean birth with a gut microbiome-based program: A randomized controlled trial
Abstract
Background: Infants born via Cesarean section (C-section) often have a distinct gut microbiome and higher risks of atopic and immune-related conditions than vaginally delivered infants. We evaluated whether a microbiome-based program could shift gut microbiome composition and improve microbiome-associated health outcomes in C-section born infants.
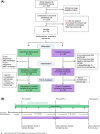
Methods: This open-label, randomized, controlled trial included full-term C-section-born infants aged 0-3 months, randomized to an intervention (n = 25) or control arm (n = 29). Over 6 months, the intervention arm received two microbiome reports, personalized recommendations based on their microbiome, educational materials, and coaching calls focused on microbiome health. Parents reported health conditions via surveys.
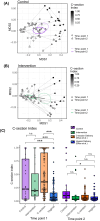
Primary outcome: Difference between study arms in relative abundance of key gut microbiome taxa and functional genes. Other outcomes: Changes in a C-section index-a taxonomy-based metric comparing C-section-associated taxa to vaginally-associated taxa-and prevalence of atopic conditions.
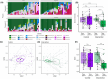
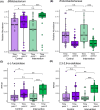
Results: Compared to controls, the intervention arm had higher Bifidobacterium (p = .025, q = .121) and higher abundance of genes associated with human milk oligosaccharide degradation (e.g., α-L-fucosidase, p = .019, q = .046) at timepoint 2. In the intervention arm, the C-section index decreased to a level similar to vaginally born infants (p = .807, q = .807). At the end of the intervention, atopic dermatitis prevalence was lower in the intervention arm than in controls (odds ratio, 0.17 [95% CI, 0.023-0.723], p = .031).
Conclusion: A personalized microbiome-based program can modulate the gut microbiome of C-section-born infants and may reduce the risk of atopic conditions (ClinicalTrials.gov: NCT06424691).
Keywords: atopic dermatitis; bifidobacterium; cesarean section; gut; infant gut; microbiome.
© 2025 Seeding Inc dba Tiny Health. Pediatric Allergy and Immunology published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
P.A.N., C.N., J.T., D.G., T.S., D.S.T., A.O., and K.V.S. are employees of Tiny Health, and except for A.O., all hold stock options in the company. K.V.S. and T.S. have been paid by Tiny Health to attend meetings. K.V.S. and D.G. have planned and pending patents related to the research. E.S. serves as the Chief Medical Officer (CMO) of Tiny Health. N.T.M. is a scientific advisor to Tiny Health. R.A.M. is a founding advisor to Tiny Health, a role approved by the Medical‐Industry Relations Committee of Mayo Clinic, fully independent of his employment at Mayo Clinic. C.S.H. is the CEO and founder of Tiny Health. Q.Y. declares no competing interests.
Figures
References
-
- Jakobsson HE, Abrahamsson TR, Jenmalm MC, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63(4):559‐566. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

