Modeling Genetic Risk of β-Cell Dysfunction in Human Induced Pluripotent Stem Cells From Patients Carrying the MTNR1B Risk Variant
- PMID: 40898610
- PMCID: PMC12405741
- DOI: 10.1111/jpi.70073
Modeling Genetic Risk of β-Cell Dysfunction in Human Induced Pluripotent Stem Cells From Patients Carrying the MTNR1B Risk Variant
Abstract
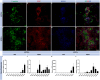
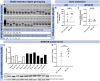
Disruptions in circadian rhythm, partly controlled by the hormone melatonin, increase the risk of type 2 diabetes (T2D). Accordingly, a variant of the gene encoding the melatonin receptor 1B (MTNR1B) is robustly associated with increased risk of T2D. This single-nucleotide polymorphism (SNP; rs10830963; G-allele) is an expression quantitative trait locus (eQTL) in human pancreatic islets, conferring increased expression of MTNR1B, which is thought to perturb pancreatic β-cell function. To understand this pathogenic mechanism in detail, we utilized human induced pluripotent stem cells (hiPSC), derived from individuals with T2D carrying the MTNR1B G-allele. Patient-derived fibroblasts were reprogrammed to hiPSC and single-base genome editing by CRISPR/Cas9 was employed to create isogenic lines of either the C/C or G/G genotypes (nonrisk and risk, respectively). In addition, the human embryonic stem cell (hESC) line (HUES4) was subjected to genome editing to create isogenic lines of either the C/C or G/G genotypes. hiPSC and hESC were differentiated into β-like cells, using a 50-day 2D protocol. Single-base genome editing generated cells with the desired genotype at a success rate of > 90%. Expression of stage-specific markers confirmed differentiation of both hiPSC and hESC into β-cells. MTNR1B mRNA levels were consistently low in differentiated β-cells, precluding quantitative analysis of gene expression. Western blot analyses indicated slightly higher levels of the MTNR1B protein in differentiated β-cells carrying the risk allele, which is in accord with the notion that rs10830963 (G-allele) functions as an eQTL in β-cells. Insulin secretion in response to the combination of high glucose and IBMX was comparable between genotypes, whereas the addition of melatonin appeared to reduce insulin secretion more efficiently in cells carrying the G-allele. While our data suggest elevated MTNR1B protein levels in stem cell-derived β-like cells carrying the risk allele, these cells do not appear to be sufficiently mature to establish rs10830963 as an eQTL at the mRNA level. The observed nominal increase in melatonin sensitivity in G-allele-carrying cells is suggestive of a functional contribution of rs10830963 to β-cell dysfunction; however, this interpretation remains tentative and will require further validation in more mature β-cell models.
Keywords: melatonin receptor 1B; single‐nucleotide polymorphism; stem cell derived β‐cells; β‐cell function.
© 2025 The Author(s). Journal of Pineal Research published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
The G-allele of rs10830963 in MTNR1B Exerts Stage-Specific Effects Across the Trajectory of Type 2 Diabetes: A Multi-State Analysis.Int J Mol Sci. 2025 Aug 14;26(16):7855. doi: 10.3390/ijms26167855. Int J Mol Sci. 2025. PMID: 40869173 Free PMC article.
-
[A ssociations of short-term ambient particulate matter exposure and MTNR1B gene with triglyceride-glucose index: A family-based study].Beijing Da Xue Xue Bao Yi Xue Ban. 2024 Jun 18;56(3):375-383. doi: 10.19723/j.issn.1671-167X.2024.03.001. Beijing Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38864120 Free PMC article. Chinese.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
The Association of rs4753426 Polymorphism in the Melatonin Receptor 1B (MTNR1B) Gene and Susceptibility to Adolescent Idiopathic Scoliosis: A Systematic Review and Meta-analysis.Pain Physician. 2015 Sep-Oct;18(5):419-31. Pain Physician. 2015. PMID: 26431121
-
Type 1 Diabetes: A Guide to Autoimmune Mechanisms for Clinicians.Diabetes Obes Metab. 2025 Aug;27 Suppl 6(Suppl 6):40-56. doi: 10.1111/dom.16460. Epub 2025 May 15. Diabetes Obes Metab. 2025. PMID: 40375390 Free PMC article. Review.
References
-
- Muoio D. M. and Newgard C. B., “Molecular and Metabolic Mechanisms of Insulin Resistance and β‐Cell Failure in Type 2 Diabetes,” Nature Reviews Molecular Cell Biology 9, no. 3 (2008): 193–205. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

