Uropathogenic Escherichia coli proliferate as a coccoid morphotype inside human host cells
- PMID: 40901850
- PMCID: PMC12407437
- DOI: 10.1371/journal.pbio.3003366
Uropathogenic Escherichia coli proliferate as a coccoid morphotype inside human host cells
Abstract
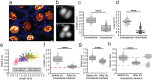
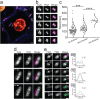
Escherichia coli is arguably one of the most studied bacterial model systems in modern biology. While E. coli are normally rod-shaped gram-negative bacteria, they are known to undergo conditional morphology changes under environmental and nutrient stress. In this study, using an infection-based in-vitro infection model system combined with advanced dynamical imaging, we present the first molecular details of uropathogenic E. coli (UPEC) dividing to form and proliferate as coccoid-shaped cells inside human host cells. For these intracellular UPEC cells, the frequency of cell division outpaced the rate of cell growth, resulting in a morphological transition from traditional rod-shape to coccobacilli. We also visualized the subcellular protein dynamics in these cells and noted that the division proteins follow the similar localization and constriction patterns that have been demonstrated for vegetative growth. However, unlike for fast-growing rod-shaped cells, FtsZ constriction in intracellular UPEC occurs prior to visual nucleoid segregation. Our results suggest that the modulation of division rate contributes to morphological adaptability of intracellular UPEC at the single-cell level.
Copyright: © 2025 Pokhrel et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

