Synaptic targets of circadian clock neurons influence core clock parameters
- PMID: 40901945
- PMCID: PMC12407047
- DOI: 10.1126/sciadv.adw4666
Synaptic targets of circadian clock neurons influence core clock parameters
Abstract
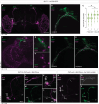
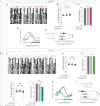
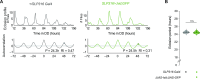
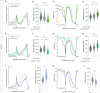
Neuronal connectivity in the circadian clock network is essential for robust endogenous timekeeping. In the Drosophila circadian clock network, the small ventral lateral neurons (sLNvs) serve as critical pacemakers. Peptidergic communication mediated by the neuropeptide Pigment Dispersing Factor (PDF), released by sLNvs, has been well characterized. In contrast, little is known about the role of the synaptic connections that sLNvs form with downstream neurons. Connectomic analyses revealed that the sLNvs form strong synaptic connections with previously uncharacterized neurons called superior lateral protocerebrum 316 (SLP316). Here, we show that silencing the synaptic output from the SLP316 neurons via tetanus toxin expression shortened the free-running period, whereas hyperexciting them by expressing the bacterial voltage-gated sodium channel resulted in period lengthening. Under light-dark cycles, silencing SLP316 neurons caused lower daytime activity and higher daytime sleep. Our results reveal that the main postsynaptic partners of key Drosophila pacemaker neurons are a nonclock neuronal cell type that regulates the timing of sleep and activity.
Figures


Update of
-
Synaptic Targets of Circadian Clock Neurons Influence Core Clock Parameters.bioRxiv [Preprint]. 2025 Feb 6:2025.01.30.635801. doi: 10.1101/2025.01.30.635801. bioRxiv. 2025. Update in: Sci Adv. 2025 Sep 5;11(36):eadw4666. doi: 10.1126/sciadv.adw4666. PMID: 39975067 Free PMC article. Updated. Preprint.
References
-
- Herzog E. D., Neurons and networks in daily rhythms. Nat. Rev. Neurosci. 8, 790–802 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

