Discovery of core genes for systemic lupus erythematosus via genome-wide aggregated trans-effects analysis
- PMID: 40903491
- PMCID: PMC12527927
- DOI: 10.1038/s41435-025-00352-4
Discovery of core genes for systemic lupus erythematosus via genome-wide aggregated trans-effects analysis
Abstract
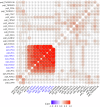
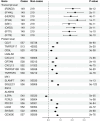
The "omnigenic" hypothesis postulates that the polygenic effects of common variants on a typical complex trait coalesce on relatively few core genes through trans-effects on their expression. Our aim was to identify core genes for systemic lupus erythematosus (SLE) by testing for association with genome-wide aggregated trans-effects (GATE) scores for gene expression in a large genetic dataset (5267/4909 SLE cases/controls). SLE was strongly associated with upregulation of expression of eight interferon-stimulated genes driven by shared trans-effects. We estimate that trans-effects on interferon signaling account for 9% of the total genetic effect on SLE risk. Outside this pathway, GATE analysis detected twenty putative core genes for SLE. Direct protein measurements for these genes were strongly associated with SLE in UK Biobank. Two putative core genes (TNFRSF17, TNFRSF13B) encode receptors (BCMA, TACI) expressed on B cells; their ligands (BAFF, APRIL) are targeted by drugs licensed or in development for SLE. Four genes (PDCD1, LAG3, TNFRSF9, CD27) encode receptors that have been characterized as immune checkpoints, and three (CD5L, SIGLEC1, CXCL13) are biomarkers of SLE disease activity. These results provide genetic support for existing drug targets in SLE (interferon signaling, BAFF/APRIL signaling) and identify other possible therapeutic targets including immune checkpoint receptors.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Mohan C, Putterman C. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat Rev Nephrol. 2015;11:329–41. - PubMed
-
- Frangou E, Vassilopoulos D, Boletis J, Boumpas DT. An emerging role of neutrophils and NETosis in chronic inflammation and fibrosis in systemic lupus erythematosus (SLE) and ANCA-associated vasculitides (AAV): Implications for the pathogenesis and treatment. Autoimmun Rev. 2019;18:751–60. - PubMed
-
- Alarcón-Segovia D, Alarcón-Riquelme ME, Cardiel MH, Caeiro F, Massardo L, Villa AR, et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheumatism. 2005;52:1138–47. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

