Rewiring of cortical glucose metabolism fuels human brain cancer growth
- PMID: 40903569
- PMCID: PMC12507665
- DOI: 10.1038/s41586-025-09460-7
Rewiring of cortical glucose metabolism fuels human brain cancer growth
Abstract
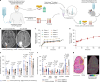
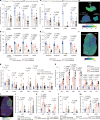
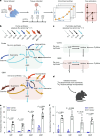
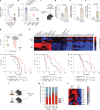
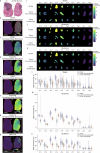
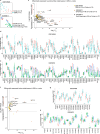
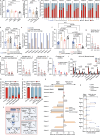
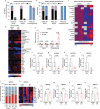
The brain avidly consumes glucose to fuel neurophysiology1. Cancers of the brain, such as glioblastoma, relinquish physiological integrity and gain the ability to proliferate and invade healthy tissue2. How brain cancers rewire glucose use to drive aggressive growth remains unclear. Here we infused 13C-labelled glucose into patients and mice with brain cancer, coupled with quantitative metabolic flux analysis, to map the fates of glucose-derived carbon in tumour versus cortex. Through direct and comprehensive measurements of carbon and nitrogen labelling in both cortex and glioma tissues, we identify profound metabolic transformations. In the human cortex, glucose carbons fuel essential physiological processes, including tricarboxylic acid cycle oxidation and neurotransmitter synthesis. Conversely, gliomas downregulate these processes and scavenge alternative carbon sources such as amino acids from the environment, repurposing glucose-derived carbons to generate molecules needed for proliferation and invasion. Targeting this metabolic rewiring in mice through dietary amino acid modulation selectively alters glioblastoma metabolism, slows tumour growth and augments the efficacy of standard-of-care treatments. These findings illuminate how aggressive brain tumours exploit glucose to suppress normal physiological activity in favour of malignant expansion and offer potential therapeutic strategies to enhance treatment outcomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.R.W. has consulted for Agios Pharmaceuticals, Admare Pharmaceuticals, Bruker and Innocrin Pharmaceuticals. D.R.W. is listed as an inventor on patents pertaining to the treatment of patients with brain tumours (US provisional patent application 63/416,146, US provisional patent application 62/744,342, US provisional patent application 62/724,337). A.J.S., D.N., C.A.L., A.M., A.A. and B.M. are listed as co-inventors on US provisional patent application 63/416,146. N.Y.R.A. reports the following disclosures: key opinion leader to Bruker Daltonics, collaboration with Thermo Finnigan, service agreement with EMD Serono, service agreement with iTeos Therapeutics, and founder and board member of BondZ. In the past three years, C.A.L. has consulted for Odyssey Therapeutics and Third Rock Ventures. W.N.A.-H. has consulted for Servier Pharmaceuticals. The other authors declare no competing interests.
Figures







Update of
-
Rewiring of cortical glucose metabolism fuels human brain cancer growth.medRxiv [Preprint]. 2023 Oct 25:2023.10.24.23297489. doi: 10.1101/2023.10.24.23297489. medRxiv. 2023. Update in: Nature. 2025 Oct;646(8084):413-422. doi: 10.1038/s41586-025-09460-7. PMID: 37961582 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 CA251872/CA/NCI NIH HHS/United States
- K99 CA300923/CA/NCI NIH HHS/United States
- U2C DK135066/DK/NIDDK NIH HHS/United States
- P50 CA269022/CA/NCI NIH HHS/United States
- R01 NS127799/NS/NINDS NIH HHS/United States
- R01 NS129123/NS/NINDS NIH HHS/United States
- R01 CA271369/CA/NCI NIH HHS/United States
- K08 CA234416/CA/NCI NIH HHS/United States
- R37 CA258346/CA/NCI NIH HHS/United States
- R35 GM143127/GM/NIGMS NIH HHS/United States
- F32 CA260735/CA/NCI NIH HHS/United States
- R01 CA261926/CA/NCI NIH HHS/United States
- R01 NS110572/NS/NINDS NIH HHS/United States
- U54 CA283114/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

