Commensal yeast promotes Salmonella Typhimurium virulence
- PMID: 40903573
- PMCID: PMC12460169
- DOI: 10.1038/s41586-025-09415-y
Commensal yeast promotes Salmonella Typhimurium virulence
Abstract
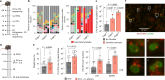
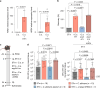
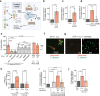
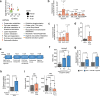
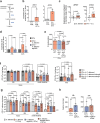
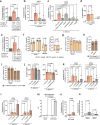
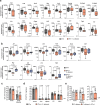
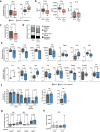
Enteric pathogens engage in complex interactions with the host and the resident microbiota to establish gut colonization1-3. Although mechanistic interactions between enteric pathogens and bacterial commensals have been extensively studied, whether and how commensal fungi affect enteric infections remain largely unknown1. Here we show that colonization with the common human gut commensal fungus Candida albicans worsened infections with the enteric pathogen Salmonella enterica subsp. enterica serovar Typhimurium. The presence of C. albicans in the mouse gut increased Salmonella caecal colonization and systemic dissemination. We investigated the underlying mechanism and found that Salmonella binds to C. albicans via type 1 fimbriae and uses its type 3 secretion system to deliver effector proteins into C. albicans. A specific effector, SopB, was sufficient to manipulate C. albicans metabolism and trigger the release of millimolar amounts of arginine into the extracellular environment. The released arginine, in turn, induced expression of the type 3 secretion system in Salmonella, increasing its invasion of epithelial cells. C. albicans deficient in arginine production was unable to increase Salmonella virulence. Arginine-producing C. albicans also dampened the inflammatory response during Salmonella infection. Arginine supplementation in the absence of C. albicans increased the systemic spread of Salmonella and decreased the inflammatory response, phenocopying the presence of C. albicans. In summary, we identified C. albicans colonization as a susceptibility factor for disseminated Salmonella infection and arginine as a central metabolite in the cross-kingdom interaction between fungi, bacteria and host.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






Update of
-
Commensal Yeast Promotes Salmonella Typhimurium Virulence.bioRxiv [Preprint]. 2024 Aug 8:2024.08.08.606421. doi: 10.1101/2024.08.08.606421. bioRxiv. 2024. Update in: Nature. 2025 Sep;645(8082):1002-1010. doi: 10.1038/s41586-025-09415-y. PMID: 39211098 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

